diff --git a/PDFs/xarray_intro_part1.pdf b/PDFs/xarray_intro_part1.pdf
new file mode 100644
index 0000000000000000000000000000000000000000..65d12f0c69fbd635d3b086be45664d1f88503ddb
Binary files /dev/null and b/PDFs/xarray_intro_part1.pdf differ
diff --git a/PDFs/xarray_intro_part2.pdf b/PDFs/xarray_intro_part2.pdf
new file mode 100644
index 0000000000000000000000000000000000000000..90b4ed6c73aedc05106f2e9aa6920c8d369c6821
Binary files /dev/null and b/PDFs/xarray_intro_part2.pdf differ
diff --git a/PDFs/xarray_introduction_part1.pdf b/PDFs/xarray_introduction_part1.pdf
deleted file mode 100644
index 07ebf104201328ce2ae121eec28d555c3122b2a5..0000000000000000000000000000000000000000
Binary files a/PDFs/xarray_introduction_part1.pdf and /dev/null differ
diff --git a/PDFs/xarray_introduction_part2.pdf b/PDFs/xarray_introduction_part2.pdf
deleted file mode 100644
index f9bb4c5abf2e4fdd688adc6c51d5951ee106b662..0000000000000000000000000000000000000000
Binary files a/PDFs/xarray_introduction_part2.pdf and /dev/null differ
diff --git a/data/hist_em_LR_temp_subset_1980-2000.nc b/data/hist_em_LR_temp_subset_1980-2000.nc
new file mode 100644
index 0000000000000000000000000000000000000000..e72559a53a99bb0cc6f920f5c04f5ec8a438dd13
Binary files /dev/null and b/data/hist_em_LR_temp_subset_1980-2000.nc differ
diff --git a/data/ssp245_em_LR_temp_subset_2070-2100.nc b/data/ssp245_em_LR_temp_subset_2070-2100.nc
new file mode 100644
index 0000000000000000000000000000000000000000..88855b40d6763e628b565af78691efe73b423921
Binary files /dev/null and b/data/ssp245_em_LR_temp_subset_2070-2100.nc differ
diff --git a/notebooks/xarray_intro_part1.ipynb b/notebooks/xarray_intro_part1.ipynb
new file mode 100644
index 0000000000000000000000000000000000000000..8f10384be66f0c0f7dd4b1d109b1e2f11b541883
--- /dev/null
+++ b/notebooks/xarray_intro_part1.ipynb
@@ -0,0 +1,2841 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "id": "08a8d53e-167a-480b-beac-15bc6b378f94",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "<p align=\"right\">\n",
+ " <img src=\"https://www.dkrz.de/@@site-logo/dkrz.svg\" width=\"12%\" align=\"right\" title=\"DKRZlogo\" hspace=\"20\">\n",
+ " <img src=\"https://wr.informatik.uni-hamburg.de/_media/logo.png\" width=\"12%\" align=\"right\" title=\"UHHLogo\">\n",
+ "</p>\n",
+ "<div style=\"font-size: 20px\" align=\"center\"><b> Python Course for Geoscientists, 5-8 March 2024</b></div>\n",
+ "<div style=\"font-size: 15px\" align=\"center\">\n",
+ " <b>see also <a href=\"https://gitlab.dkrz.de/pythoncourse/material\">https://gitlab.dkrz.de/pythoncourse/material</a></b>\n",
+ "</div>\n",
+ "\n",
+ "***"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "7128b107-2215-4d84-a411-8e0e42fb9a87",
+ "metadata": {},
+ "source": [
+ "\n",
+ "<p align=\"center\">\n",
+ " <img src=\"https://docs.xarray.dev/en/stable/_static/Xarray_Logo_RGB_Final.svg\" width=\"35%\" align=\"right\" title=\"xarraylogo\" hspace=\"20\">\n",
+ "</p>\n",
+ "\n",
+ "<font size=\"20\"> xarray introduction I</font> \n",
+ "\n",
+ "xarray documentation: https://docs.xarray.dev/en/stable/index.html\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e81e31bd-f599-448f-93b8-8017e91133ae",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "slide"
+ },
+ "tags": []
+ },
+ "source": [
+ "## What is xarray?\n",
+ "\n",
+ "\n",
+ "*xarray* is a Python package that simplifies the handling of *multi-dimensional datasets*. It offers a wide range of functions for advanced analytics and visualization, expanding upon the capabilities of *NumPy, Pandas, and Matplotlib.*\n",
+ "\n",
+ "The underlying data model of xarray is based on the [netCDF data format](http://www.unidata.ucar.edu/software/netCDF). This format, along with adherence to the [Climate and Forecast metadata conventions](https://cfconventions.org/) (CF), is standard in the climate science community. As a result, xarray enables *efficient and intuitive data analysis* on such datasets. However, it also supports other file formats like *GRIB, HDF5, and Zarr*."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6e5188ab",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "slide"
+ },
+ "tags": []
+ },
+ "source": [
+ "## xarray\"s Data Structure\n",
+ "see also https://tutorial.xarray.dev/fundamentals/01_datastructures.html\n",
+ "\n",
+ "**DataArray**: \n",
+ "A DataArray in xarray is essentially a NumPy array enriched with additional metadata such as dimension names, coordinates, and attributes. It serves as a fundamental building block, akin to a data variable object.\n",
+ "\n",
+ "**Dimensions**: \n",
+ "Dimensions represent the axes along which the data is organized. In xarray, each dimension is assigned a unique name.\n",
+ "_Note: Commonly, the dimension order in multidimensional Earth Science data is (time, level, latitude, longitude)_\n",
+ "\n",
+ "**Coordinates**: \n",
+ "Coordinates provide descriptive labels for the dimensions of your data, offering additional context and meaning to the data points along each dimension.\n",
+ "\n",
+ "**Attributes**: \n",
+ "Attributes, also known as metadata, are additional pieces of information attached to DataArrays, coordinate variables and/or Datasets. They include descriptive information such as units, CF standard_name, FillValue attributes, and comments.\n",
+ "\n",
+ "**Dataset**: \n",
+ "A Dataset in xarray is a dictionary-like collection of one or more DataArray objects with aligned dimensions. It serves as a container for organizing and managing multiple data variables within a coherent structure, mirroring the structure of a netCDF data file object.\n",
+ "\n",
+ "----\n",
+ "\n",
+ "<br>\n",
+ "\n",
+ "<img src=\"https://storage.googleapis.com/jnl-up-j-jors-files/journals/1/articles/148/submission/proof/148-10-1829-1-17-20170405.png\" alt=\"xarray data structure\" border=1 width=900></img> \n",
+ "<figcaption align = \"center\"> An overview of xarray’s main data structures. From Hoyer and Hamman (2017); DOI: 10.5334/jors.148 </figcaption> <br>"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "252d06c6-fa7c-46e3-8241-2c17d50d856d",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "<br>\n",
+ "\n",
+ "========================================================================================\n",
+ "# Importing Modules and Configure the Notebook\n",
+ "========================================================================================"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "77c14150-5cc1-42c2-b819-c0351ba9edfb",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "import xarray as xr\n",
+ "import numpy as np\n",
+ "import pandas as pd\n",
+ "try:\n",
+ " import cfgrib\n",
+ "except ImportError:\n",
+ " import subprocess\n",
+ " subprocess.run([\"bash\", \"-c\", \"pip install --user ecmwflibs --quiet\"])\n",
+ "\n",
+ "from datetime import datetime\n",
+ "import matplotlib.pyplot as plt\n",
+ "import matplotlib as mpl\n",
+ "\n",
+ "\n",
+ "# Set this to render all evaluated output of a cell\n",
+ "from IPython.core.interactiveshell import InteractiveShell\n",
+ "InteractiveShell.ast_node_interactivity = \"all\"\n",
+ "\n",
+ "\n",
+ "# Set default figure size and font size\n",
+ "mpl.rcParams.update({\n",
+ " \"figure.figsize\": (3.5, 2.5),\n",
+ " \"font.size\": 9\n",
+ "})"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "b87bed17-e651-4791-850e-688c75afb20c",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ }
+ },
+ "source": [
+ "<br>\n",
+ "\n",
+ "======================================================================\n",
+ "# Showcases and Exercises\n",
+ "======================================================================\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f1c5892f-918b-4c2e-8880-df278c6485bb",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "## _(A) xarray DataArrays_\n",
+ "***\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "7f87cdb2-6255-4e18-8299-08e004b96ded",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "The `xr.DataArray()` function in xarray is used to create a multi-dimensional array-like object that encapsulates a NumPy array or another array-like data structure (referred to as data). This function accepts optional additional keyword arguments to provide **labeled dimensions (dims), coordinates (coords), a name (name), and metadata (attrs)**. Below is the general syntax:\n",
+ "\n",
+ "```python\n",
+ "xr.DataArray(data,\n",
+ " coords=None,\n",
+ " dims=None,\n",
+ " name=None,\n",
+ " attrs=None\n",
+ " )\n",
+ "```\n",
+ "\n",
+ "The function `xr.DataArray()` returns a data object with a default data structure, consisting of dimension names, coordinates, indexes, and attributes. *If not provided with the `xr.DataArray()` function call, these presettings are empty.* The dimensions have the default names dim_0, dim_1,.... You can, however, modify them afterwards. \n",
+ "\n",
+ "It\"s important to **configure coordinate values** properly not only for xarray but also for other software tools. **Labeled geospatial** information from coordinates is crucial for various tasks, including:\n",
+ "\n",
+ "* **Plotting**: Mapping data onto a real-world grid for visualization.\n",
+ "* **Analysis**: Performing routines such as calculating area-weighted means.\n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "62bb81bf-7e82-4f47-a2ea-7a7f70e33ed5",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 1: Construct a \"plain\" xarray DataArray and Modify it"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "bfd34065-71cd-4a28-be7b-41fb9d26f14b",
+ "metadata": {},
+ "source": [
+ "To create a basic xarray DataArray, you can simply pass a NumPy array to the `xr.DataArray()` function call. \n",
+ "__Note__: *xr* serves as an alias for the *imported xarray package*.\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "3744f16b-3b33-4fd2-a308-45ec4e121db4",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Creating a NumPy array with shape (4, 5).\n",
+ "nlat, nlon = 4, 5\n",
+ "ndarray = np.random.rand(nlat, nlon) * 10 # or np.arange(1, nlat * nlon + 1).reshape(nlat,nlon) \n",
+ "ndarray"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "509692e3-ae14-4f54-a700-e51eea48c0f7",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Creating a plain _xarray_ DataArray.\n",
+ "da1 = xr.DataArray(ndarray)\n",
+ "da1"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f9bbb85d-55c4-4807-bf51-0e43fadcb18f",
+ "metadata": {},
+ "source": []
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "cd34960e-2b71-4620-ac7c-1d5322b332af",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Renaming dimension names of the DataArray with .rename().\n",
+ "\n",
+ "# Note: this function creates a new DataArray object, it does not modify da1 in-place.\n",
+ "da2 = da1.rename({\"dim_0\": \"lat\", \"dim_1\": \"lon\"})\n",
+ "da2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "06c9b6d7-4902-454e-aaf6-ea8c0bd3f260",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Creating coordinate variables and assigning them to the DataArray da2.\n",
+ "lons = np.linspace(0., 20., nlon)\n",
+ "lats = np.linspace(-3., 3., nlat)\n",
+ "# print(lons, lats)\n",
+ "# Note: this function creates a new DataArray object, it does not modify da2 in-place.\n",
+ "da3 = da2.assign_coords({\"lon\": lons, \"lat\": lats})\n",
+ "da3"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "3da5057c-5a49-4db1-80b5-d2a68b13c370",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Assigning a name to the DataArray da3.\n",
+ "da3.name = \"var1\"\n",
+ "da3"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "30ccdbba-5860-42e0-b9af-77da0f5d26ee",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Attach metadata attributes to the data array.\n",
+ "#== For CF standard names, see https://cfconventions.org/Data/cf-standard-names/current/build/cf-standard-name-table.html\n",
+ "da3.attrs[\"standard_name\"] = \"age_of_sea_ice\"\n",
+ "da3.attrs[\"units\"] = \"mm\""
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e191937e-9571-49c9-8ed3-9dc85357b61f",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "da3"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e1836b8e-3827-4cc8-967e-60a77c02c439",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "_**Note**_: There are two options to access data variables and coordinate variables:\n",
+ " * da3.MYVAR: attribute-style access\n",
+ " * da3[\"MYVAR\"]: dictionary-style"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "286d9f25-9854-41ad-af9d-4c4f9b3dc967",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Attach metadata attributes to the lon coordinate of the DataArray.\n",
+ "\n",
+ "da3.lon.attrs[\"standard_name\"] = \"longitude\" #- same as da3[\"lon\"].attrs[\"standard_name\"] = \"longitude\"\n",
+ "da3[\"lon\"].attrs[\"units\"] = \"degrees_east\"\n",
+ "# Note: You can also attach these attributes in a single command with a dictionary (see da4)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f7368d01-ec89-4e49-adfc-794798ba3487",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "da3"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "81cb4894-cf31-45cb-b6d2-0a18fb0014ad",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 2: Create a fully specified DataArray with all the metadata attached"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e3e071e7-e5f4-40ea-a1ac-c79bfc171c51",
+ "metadata": {},
+ "source": [
+ "When creating a DataArray with xr.DataArray(), you can directly include:\n",
+ "\n",
+ "* name: Assign a name to the DataArray.\n",
+ "* dims: Specify dimension names (as tuple)\n",
+ "* coords: Define dimension values for alignment and indexing \n",
+ " (key=>dimension name; value=> array/list representing coordinate values)\n",
+ "* attrs: Attach metadata attributes to the DataArray (as dictionary)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "8a1da962-beb0-4ac3-97cf-fdac4568bd9c",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "da4 = xr.DataArray(data=ndarray, \n",
+ " name=\"var1\",\n",
+ " dims=(\"lat\",\"lon\"), \n",
+ " coords={\"lat\": lats, \n",
+ " \"lon\": lons},\n",
+ " attrs={\"standard_name\":\"age_of_sea_ice\",\n",
+ " \"units\": \"mm\"})"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "5d0fdc2f-c8c4-4486-95f3-71a81eb7366d",
+ "metadata": {},
+ "source": [
+ "Note: The xr.DataArray() function does not allow for direct specification of coordinate metadata within the `coords`\u001b parameter. These metadata attributes for coordinates have to be added afterwards."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "95786b3a-9039-455d-9504-0341ca7da3b1",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "da4[\"lon\"].attrs={\"standard_name\":\"longitude\", \"units\":\"degrees_east\"}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2cbd0c1d-f75c-4838-b451-03d9415c8477",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "da4"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "b2b89079-5405-414c-9f6e-94d7a120937c",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 3: Plotting functionality on xarray objects "
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6b0cb7af-b1d8-4fd2-84ff-3372ccbd4da6",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "xarray DataArray objects can be plotted by invoking the `plot()` function. <br> \n",
+ "\n",
+ "Principally, it is possible to customize the plot by providing arguments. <br>\n",
+ "However, compared to more advanced plotting libraries like Matplotlib, there is only *a limited set of customization options*."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "c0731464-5fc3-4a85-a159-231372c43e5a",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "When plotting xarray DataArray objects using the `plot()` function, the default chart type is automatically determined based on the dimensionality of the data. <br>\n",
+ "You can also explicitly specify a chart type by using `plot.CHARTTYPE()`, with CHARTTYPE being line, hist, pcolormesh, scatter, etc...<br> \n",
+ "The default chart types are: \n",
+ "- 1-D data: Line2D plot                     ==> equivalent to plot.line()\n",
+ "- 2-D data: QuadMesh plot (resembling a filled contour plot)    ==> equivalent to plot.pcolormesh()\n",
+ "\n",
+ "see https://docs.xarray.dev/en/stable/generated/xarray.DataArray.plot.html\n",
+ "\n",
+ "_**NOTE**_:By default, the `plot()` function uses metadata provided by the DataArray for plotting the labels.\n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "fac7454e-77df-4521-8242-23d083079849",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Plot da4 with some customization options.\n",
+ "da4.plot(figsize=(3, 2), cmap=\"Reds\", add_colorbar=True,\n",
+ " cbar_kwargs={\"label\": \"This is the colorbar label\",\n",
+ " \"shrink\": 1.1})\n",
+ "\n",
+ "#== Additional customizations using Matplotlib functions \n",
+ "#== since these are not accessible with the plot() function.\n",
+ "plt.title(\"mytitle\");\n",
+ "plt.ylabel(\"latitude\");\n",
+ "plt.grid(\"on\");"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6c2d732e-99c6-4b31-a906-ba23b5c4bc20",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Create a 1D DataArray by slicing and plot a line plot\n",
+ "da4[:,0].plot(figsize=(3, 2));"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6e4dd3a2-5278-4ba9-a6e7-a01afae88cd5",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== You can also specify the targeted plot type function (e.g. for histogram or scatter).\n",
+ "da4.plot.hist(figsize=(3, 2)); da4.plot.scatter(figsize=(3, 2),x=\"lon\"); "
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e2e98687-dfc5-498e-988f-0a6183f5fb4b",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "slide"
+ },
+ "tags": []
+ },
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 4: From NumPy arrays to xarray DataArrays"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "0ef4c102-44df-4fa6-b0ff-222f8db56ae8",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "slide"
+ },
+ "tags": []
+ },
+ "source": [
+ "You can directly convert a NumPy array into an xarray DataArray type by using it as input for xarray\"s function `xr.DataArray`. "
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "0ef04f84-eb97-4476-83e8-98e7a4d83ede",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "slide"
+ },
+ "tags": []
+ },
+ "source": [
+ "We use the _atmosphere water vapor content_ data from the file `../data/prw.dat` by loading it with NumPy."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "56fb99e3",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Show the first 5 lines of the ascii input file.\n",
+ "!head -5 ../data/prw.dat"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "a21e702a-ad00-4b37-a0fc-9396c9e831f5",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Read columns 1 to 3 of the input file while skipping the header.\n",
+ "prw_data = np.loadtxt(\"../data/prw.dat\", usecols=(1,2,3),skiprows=1) #-- data are read in as float64\n",
+ "prw_stations = np.loadtxt(\"../data/prw.dat\", usecols=0, skiprows=1, dtype=\"U10\") #-- data are read in as string"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "169713cf-7534-412a-9dbe-689273cc2ef4",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Get the shape of the NumPy ndarray prw_data.\n",
+ "prw_data.shape"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "0bb6b812-310d-4fcf-8bc7-f7e80d04349e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Get the first 4 rows of the prw_data.\n",
+ "prw_data[:4, :]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "c0e6134e-fdd4-4433-8541-e6ddf14bbe88",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Convert the NumPy ndarray prw_data into an xarray DataArray.\n",
+ "prw_da1 = xr.DataArray(prw_data)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "dff469f0-9b16-4de3-9bc6-1359b2dead4e",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Get the first four elements (rows) of the xarray DataArray using slicing.\n",
+ "prw_da1[:4, :]"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "447993da-7487-4ed0-b07b-353ad5044da2",
+ "metadata": {},
+ "source": [
+ "The comparison of `prw_da1[:4, :])` and `prw_data[:4, :]` illustrates that when slicing a xarray DataArray,the structure and metadata are preserved. In contrast, slicing a NumPy array results in a new NumPy array without additional metadata."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "a3b2fbc0-e14a-4a39-b2f3-341ebda4a564",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# As an alternative to prw_da1[:4, :], you can use the functionality `head` on the xarray DataArray.\n",
+ "# This functionality is not available for a NumPy ndarray.\n",
+ "# prw_da1.head(4)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "624a60e6-b2a9-4612-bcfd-60dcdc3f22a5",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "print(f\"type of prw_data: {type(prw_data)}; type of prw_da1: {type(prw_da1)};\")\n",
+ "print(f\"the data structure wrapped in the xarray DataArray is: {type(prw_da1.data)}\") \n",
+ "print(f\"the data type of the elements of the wrapped data is: {prw_da1.data.dtype}\") \n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "b9ece729-9e8d-4c97-8380-0e60183851f8",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ }
+ },
+ "source": [
+ "Unlike a NumPy array, the xarray DataArray can differentiate the variable of interest as a *data variable* (`prw_da1.data`) from *coordinate* variables. This is because the xarray DataArray has the following components:\n",
+ "- **dimensions**: Named dimensions that define the structure of the array (`prw_da1.dims`).\n",
+ "- **coordinates**: Variables associated with each dimension, providing context and labeling (`prw_da1.coords`).\n",
+ "- **attributes**: Additional metadata describing the array or its components (`prw_da1.attrs`).\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "c18c8d09-e4f1-4ca5-a7cf-57705c8d2b59",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#print(f\"data variable: {prw_da1.data}\")\n",
+ "print(f\"coordinate variable: {prw_da1.coords}\") \n",
+ "print(f\"dimension names: {prw_da1.dims}\")\n",
+ "print(f\"data variable name: {prw_da1.name}\")\n",
+ "print(f\"data variable metadata: {prw_da1.attrs}\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "2a4dfdc9-b5bc-4b99-a773-6287f3a7a161",
+ "metadata": {},
+ "source": [
+ "When created without specific parameters in the xr.DataArray function call, prw_da1 defaults to empty coordinates, default dimension names (\"dim_0\", \"dim_1\"), and lacks a data variable name or associated metadata."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "55a4776e-800d-418a-adbf-89fe19e9b437",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "slide"
+ }
+ },
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 5: Specify the structure/parameters during the `xarray.DataArray()` function call"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "4354811d-84ff-42ea-be8f-046d0d692a1a",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "slide"
+ }
+ },
+ "source": [
+ "Create a new xarray DataArray called prw_da2:\n",
+ "1. The **data variable** corresponds to the first column of the NumPy array `prw_data`.\n",
+ "2. We have one dimension (**dims**) which represents individual stations; we name it **_Station_**. <br>\n",
+ " By default, it is an index running from 0 to the length of a column minus 1.\n",
+ "3. The **coords** are the first and second columns of the NumPy array `prw_data`. We want to call them `lat` and `lon`. <br>\n",
+ " They have the same dimensions as the data array, namely **_Station_**. \n",
+ " The general syntax for declaring the coordinate is: <br>\n",
+ " `coords = {coords_name: (dimension_name, coordinate_values)}`\n",
+ "4. The **name** of the data variable is **prw**.\n",
+ "5. In the **attrs**, we can store variable attributes like **_units_**. The **standard_name** of prw is **_atmosphere_mass_content_of_water_vapor_**; <br> the corresponding canonical units are **_kg m-2_**."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6f65ead5-64a1-48c4-b981-7cf253a7c748",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "prw_da2 = xr.DataArray(prw_data[:,2],\n",
+ " dims=[\"Station\"],\n",
+ " coords={\"lat\":(\"Station\", prw_data[:,0]),\n",
+ " \"lon\":(\"Station\", prw_data[:,1])},\n",
+ " name=\"prw\",\n",
+ " attrs={\"units\":\"kg m-2\",\n",
+ " \"standard_name\":\"atmosphere_mass_content_of_water_vapor\"})"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "9831c927-346e-4f1d-80d0-0f910be7856d",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "prw_da2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "588b23fa-9c50-40bc-9a58-2dd4d50cb7fa",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "print(f\"data variable (sliced): {prw_da2.data[:8]}\")\n",
+ "print(f\"dimension names: {prw_da2.dims}\")\n",
+ "\n",
+ "print(f\"coordinate variable:\\n {prw_da2.coords}\") \n",
+ "\n",
+ "print(f\"name of DataArray (coresponding to data variable name): {prw_da2.name}\")\n",
+ "print(f\"metadata of DataArray (corresponding to data variable metadata):\\n {prw_da2.attrs}\")\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "39ea8ad8-345f-4b7c-84ea-d85f1b257918",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "print(f\"shape the DataArray: {prw_da2.shape}\")\n",
+ "print(f\"size of the DataArray: {prw_da2.sizes}\") \n",
+ "# Note: sizes returns a eturns an immutable dictionary-like object, often referred to as a \"frozen\" object in xarray."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6689de19-4091-40d7-b32e-13e7fcf7f189",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "prw_da2.plot(figsize=(3, 2));"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "a3fe1395-c020-4203-a77d-27839a2ec49d",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Exercise 1: `xr.DataArray`\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "ed37edda-48b4-4447-954f-e0779b19dd11",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 1. Generate a 3-dimensional xarray DataArray.\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "bcd00fc9-eec6-4341-baf4-5b1fa5ea66d6",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 2. Add some attributes, including a standard_name attribute.\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "04d2be73-0c53-4cd9-929d-ebd0a3a153d9",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 3. Change the default dimension names and add coordinate values. \n",
+ "# (Hint: use .assign_coords(t=tcoords, y=ycoords, x=xcoords)\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "b3492698-e9e4-460a-bc2a-29c13dbf320d",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 4. Create the same DataArray with just one call of xr.DataArray.\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "b3bd0f5b-8e7e-4aed-963d-fc36154430ac",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "<br>\n",
+ "\n",
+ "### Solution Exercise 1"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6783227d-9eda-4d61-98ab-a429873fe48a",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 1.\n",
+ "ntime, nlat, nlon = 5, 6, 2\n",
+ "mydata = np.random.random((ntime, nlat, nlon))\n",
+ "data_xr = xr.DataArray(mydata)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "a4fffa91-9d40-4762-bf68-ca978cc069a2",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "data_xr.head(2)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "10c8e399-6de1-40c9-a8b0-b72237e477ee",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 2.\n",
+ "data_xr.attrs={\"standard_name\": \"fire_temperature\", \"units\": \"K\", \"my_attr\": \"my new attribute 1\"}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "441cfcba-0668-424a-9183-ed322f47094d",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 3. \n",
+ "data_xr1 = data_xr.rename({\"dim_0\":\"time\", \"dim_1\":\"lat\", \"dim_2\":\"lon\"})\n",
+ "tcoords = pd.date_range(\"2022-03-01\", periods=ntime, freq=\"D\")\n",
+ "ycoords = [30, 31, 32, 33, 34, \"b\"]\n",
+ "xcoords = np.linspace(1, 4, nlon)\n",
+ "data_xr2 = data_xr1.assign_coords(time=tcoords, lat=ycoords, lon=xcoords)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "b5cd5267-374b-471e-b927-757904d60a89",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "data_xr2.head(2)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "59c1b03d-b2bc-482d-a86e-58e4597ded2d",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 4. Create the same DataArray with just one call of xr.DataArray.\n",
+ "data_xr3 = xr.DataArray(mydata, \n",
+ " dims=(\"time\", \"lat\", \"lon\"), \n",
+ " coords={\"time\": tcoords, \n",
+ " \"lat\": ycoords,\n",
+ " \"lon\": xcoords},\n",
+ " attrs={\"standard_name\":\"fire_temperature\",\n",
+ " \"units\":\"K\", \n",
+ " \"my_attr\": \"my new attribute 1\"})\n",
+ "\n",
+ "data_xr3.head(2)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "1f9d1a71-cfec-4402-942e-27dde0d58102",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "slide"
+ },
+ "tags": []
+ },
+ "source": [
+ "***\n",
+ "\n",
+ "### Exercise 2: `xr.DataArray`\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e0717073",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ }
+ },
+ "outputs": [],
+ "source": [
+ "# 1. Create a two dimensional NumPy called prw_data_2d with the size `len(prw_data)` x `len(prw_data)` \n",
+ "# and assign `NaN` values to the entire array.\n",
+ "# Use np.Nan and np.full() or np.empty().\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "4baf6447-377d-4abb-88ac-4dbc55efa73c",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 2. On the diagonal of the quadratic array, insert the values of prw_data. Use a for loop.\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "28e4d302-85ab-4d2f-a67c-0b0e474e4d96",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 3. Convert the NumPy array to a xarray DataArray named prw_da3.\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "34840ed8-55d0-40ab-9e43-08cb415216cf",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 4. Show prw_da3 by plotting.\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f75d2261-bf09-4f48-b3fa-4ebe004a5b05",
+ "metadata": {},
+ "source": [
+ "### Solution Exercise 2"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2dbcd161-a5a2-4d98-a6fd-17bfb44ec015",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 1.\n",
+ "#prw_data.shape\n",
+ "prw_data_2d = np.full([len(prw_data), len(prw_data)], np.nan) #-- np.full creates an array with a specified value\n",
+ "# Alternatives:\n",
+ "# prw_data_2d = np.empty([len(prw_data),len(prw_data)]) * np.nan"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "daf21374-4bb0-4305-9479-853d90b422f3",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 2. \n",
+ "for i in range(0, len(prw_data)):\n",
+ " prw_data_2d[i,i] = prw_data[i,2]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "beb4c823-b603-4232-b4c6-cdf8cdce5533",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 3. \n",
+ "prw_da3 = xr.DataArray(prw_data_2d)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "b97580ea-645d-4d83-924d-0d43dfce82b5",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 4.\n",
+ "prw_da3.plot();"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f75cb937-b30d-4270-82ff-f599ecaecad7",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ }
+ },
+ "source": [
+ "### Showcase 6: Gridding station data with `xr.DataArray`\n",
+ "\n",
+ "\n",
+ "To put the station data prw_data from showcase 1 on a regular lat lon grid, use the following steps: "
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "fb50f9bb-2f3a-481f-bf84-71b557dc7126",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ },
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== 1. Define the latitudes and longitudes for the regular grid using np.linspace, \n",
+ "# with the minimum and maximum values of latitude and longitude from the station coordinates as bounds.\n",
+ "\n",
+ "nlat, nlon = 25, 50\n",
+ "latitudes = np.linspace(min(prw_data[:,0]), max(prw_data[:,0]), num=nlat)\n",
+ "longitudes = np.linspace(min(prw_data[:,1]), max(prw_data[:,1]), num=nlon)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "8df7b816-5ca3-4d6a-ae28-1589f18790cd",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ },
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== 2. Create a 2D grid of NaN values with the size (nlat,nlon).\n",
+ "data_grid = np.full((nlat, nlon), np.nan)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "adb6c672-dba4-49ec-b203-19c2a393fa8d",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ },
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== 3. Find the indices for assigning data values to the grid.\n",
+ "lat_indices = np.searchsorted(latitudes, prw_data[:,0])\n",
+ "lon_indices = np.searchsorted(longitudes, prw_data[:,1])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "cc4f13b1-52bd-4714-8dd5-c0cbaea621d4",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== 4. Assign the data values to the corresponding grid points.\n",
+ "data_grid[lat_indices, lon_indices] = prw_data[:,2]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "5455c807-8de5-48f7-b63c-ef9f3dc836c2",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== 5. Create the DataArray with latitude and longitude coordinates.\n",
+ "pwr_da4 = xr.DataArray(data_grid, \n",
+ " coords={\"lat\": latitudes, \"lon\": longitudes}, \n",
+ " dims=[\"lat\", \"lon\"])"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "bfbc320a-5149-407a-9bf6-e0f242c98a93",
+ "metadata": {},
+ "source": [
+ "6. Plot the new DataArray"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "500ac270-0880-4e28-9a4c-8ee349e04a6f",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "pwr_da4.plot();"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "dbb9eef5-45eb-4c30-8315-63928791efa9",
+ "metadata": {},
+ "source": [
+ "### Showcase 7: Combine of xarray plotting with more advanced matplotlib/cartopy features for creating maps"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "363b1960-5daa-4fa4-9de9-329abf64a0c5",
+ "metadata": {},
+ "source": [
+ "In this showcase, we use the regularly gridded station data pwr_da4 from Showcase 6."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "50700c8c-a189-4ca3-9438-b123813c3321",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "skip"
+ },
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "import cartopy.crs as ccrs\n",
+ "proj = ccrs.PlateCarree() # choose map projection\n",
+ "fig, ax = plt.subplots(figsize=(7, 9), subplot_kw={\"projection\":proj})\n",
+ "ax.set_extent([-102, -90, 29, 41], proj)\n",
+ "img = ax.stock_img() # add satellite image as background\n",
+ "img.set_alpha(0.4) # adjust background image transparency\n",
+ "ax.gridlines(draw_labels=True, color=\"None\", zorder=0) # turn on axis label, turn off gridlines\n",
+ "ax.coastlines() # add coastlines\n",
+ "pwr_da4.plot(cmap=\"turbo\", cbar_kwargs={\"shrink\": 0.5, \"pad\": 0.1});"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "1956f70a",
+ "metadata": {
+ "slideshow": {
+ "slide_type": "subslide"
+ }
+ },
+ "source": [
+ "<br>\n",
+ "\n",
+ "***\n",
+ "\n",
+ "### Exercise 3: `xr.DataArray` Modify pwr_da4\n",
+ "\n",
+ "<br>"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "d8de744d-f91c-4d20-b3b5-96baba2facf2",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 1. print the variable attributes and add a variable long_name attribute \n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "56cc7d97-ee01-4bbf-a0c6-65f1171574b4",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 2. print the variable name and change it\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6ebad36e-a623-43ca-a8dd-c8316f1a84db",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 3. print the dimension names.\n",
+ "# rename them into lat1 and lon and assign the returned object as pwr_da5\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "15c68af8-08cb-4cd5-89c6-c716140b800f",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 4. add a standard_name and units attribute to the coordinate variable lon1 in pwr_da5\n",
+ "# and print the new attributes\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "7585ab2f-97c6-4ef5-a7a5-bc1f182965d4",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 5. exchange the lat1 coordinate variable values by a numpy array;\n",
+ "# print the first 5 values of the new lat1 coordinate variable \n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "3b92e684-221c-441d-86d6-28306e58e491",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 6. set the values of the first 6 rows and first 6 columns of \n",
+ "# the pwr_data_da4 DataArray to -50\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "80b66dc3-905e-45ed-8adf-481f0bb9e4b2",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 7. plot the modified DataArray \n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "da08ff6e-4cf4-4ee1-b67f-d656eb68e123",
+ "metadata": {},
+ "source": [
+ "### Solution Exercise 3"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "7f3ba964-0336-4001-9c8c-d67acd4bc1e5",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 1.\n",
+ "print(f\"variable attributes is: {pwr_da4.attrs}\")\n",
+ "pwr_da4.attrs={\"long_name\": \"water vapor content\"}"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f4bf07af-ad05-4c6c-9b86-6fc3e32e599a",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 2.\n",
+ "print(f\"variable name is: {pwr_da4.name}\")\n",
+ "pwr_da4.name = \"prw\""
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "634519ad-778b-41fe-836d-41fa1b05e72a",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 3.\n",
+ "print(f\"dimension names are: {pwr_da4.dims}\")\n",
+ "pwr_da5 = pwr_da4.rename({\"lat\": \"lat1\", \"lon\": \"lon1\"})\n",
+ "print(f\"dimension names are: {pwr_da5.dims}\")\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "7db4bf3e-a677-4bce-9e9b-3b9f59cec5a1",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 4.\n",
+ "pwr_da5[\"lon1\"].attrs={\"standard_name\": \"longitude\", \"units\":\"degrees_east\"}\n",
+ "pwr_da5.lon1.attrs"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "666f635d-e91d-45da-8c2e-5e352fd4a35a",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 5. \n",
+ "newlats = np.linspace(0,10, pwr_da5.sizes[\"lat1\"])\n",
+ "pwr_da5.coords[\"lat1\"] = newlats\n",
+ "print(f\"first 5 values of new lat1 coordinate variable are: {pwr_da5.lat1.head(5).values}\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "36e9732f-6e09-49ae-aecd-47be33c2485b",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 6. \n",
+ "pwr_da5[:6, :6] = -50"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "b3e77203-d69e-4b36-9f6c-52bbad99310e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 7.\n",
+ "pwr_da5.plot();"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "a0ba3fef-1a8f-4dab-8d49-f64d9cef9920",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "## _(B) xarray Datasets_\n",
+ "***\n",
+ "<br>"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "2cb88b5d-fa6f-4517-9c8e-f8f0274d7535",
+ "metadata": {},
+ "source": [
+ "\n",
+ "An xarray `Dataset` is a dictionary-like container of data arrays with aligned dimensions. <br><br>\n",
+ "\n",
+ "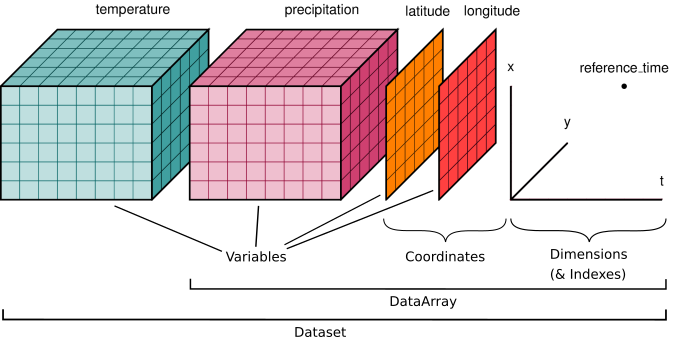\n",
+ "\n",
+ "Datasets have four key properties:\n",
+ "\n",
+ " 1. dims: dict for dimension names\n",
+ " 2. data_vars: dict of data arrays\n",
+ " 3. coords: dict of coordinates\n",
+ " 4. attrs: dict for Dataset (global) attributes\n",
+ "\n",
+ "**Note:** <br>\n",
+ "If you are familiar with the **netCDF file format**: the xarray Dataset is designed as an in-memory representation of the netCDF data model.\n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "4ad1ddf4-041a-499f-a41d-68ee88ff147e",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 8: Construct an xarray Dataset from an xarray DataArray"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "d0ec789f-0ee7-4cb1-b668-167616e7a899",
+ "metadata": {},
+ "source": [
+ "You can transform an xarray DataArrays into a xarray Dataset by using the DataArray.to_dataset() function call."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "729203d4-5d77-45b9-bc20-25a81277567c",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== We use the xarray DataArray from showcase 7.\n",
+ "ds1 = prw_da2.to_dataset()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "d665a100-55b9-4abe-885a-a5a0f0a1bc5f",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Print the xarray Dataset.\n",
+ "ds1"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "5a4e7573-ac8d-4e51-b7eb-24bd4a7651d8",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 9: Construct an xarray Dataset with the xr.Dataset() function call"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e237fb2e-4722-4f6e-a683-47255f1a22e1",
+ "metadata": {},
+ "source": [
+ "<br>\n",
+ "We here use the 3D NumPy array named `mydata` created in solution Exercise 1. <br>\n",
+ "We also use the Numpy arrays for ycoords, ycoords, tcoords created in solution Exercise 1.<br>\n",
+ "In this showcase, we create the Dataset without prior creation of a xarray DataArray."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f09136ea-d1cc-406f-ac41-3a7532f92c86",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Create an xarray Dataset with two DataArrays named \"temp\" and \"precip\". \n",
+ "#== Both DataArrays (i.e. data variables) share the same dimensions. \n",
+ "#== In the following, we show the Dataset creation into several steps:\n",
+ "#== First, we create the data variables; the syntax is:\n",
+ "\n",
+ "# data_vars = {\n",
+ "# \"var_name_1\": (dimensions_1, data_1 [, attrs_1]),\n",
+ "# \"var_name_2\": (dimensions_2, data_2 [, attrs_2]),\n",
+ "# ...}\n",
+ "data_vars = {\"temp\": ([\"time\",\"lat\",\"lon\"], mydata, {\"units\":\"K\"}),\n",
+ " \"prec\": ([\"time\",\"lat\",\"lon\"], np.square(mydata))}\n",
+ "\n",
+ "#Alternatively, you could explicitly create an xarray DataArrays:\n",
+ "#data_vars = {\"temp\": xr.DataArray(mydata, dims=[\"time\",\"lat\",\"lon\"], \n",
+ "# attrs={\"units\": \"K\", \"standard_name\": \"surface_temperature\"}),\n",
+ "# \"prec\": xr.DataArray(np.square(mydata), dims=[\"time\",\"lat\",\"lon\"]),}\n",
+ "\n",
+ "# Secondly, we define the coordinates by passing NumPy arrays\n",
+ "coords = {\"time\": tcoords,\n",
+ " \"lat\": ycoords,\n",
+ " \"lon\": xcoords}\n",
+ "# Thirdly, we define the global attributes\n",
+ "attrs = {\"history\": \"created on YYYY-MM-DD\",\n",
+ " \"creator\": \"AH\",}\n",
+ "# Create the Dataset with data variables, coordinates, and dimensions\n",
+ "ds2 = xr.Dataset(data_vars=data_vars, coords=coords, attrs=attrs)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2a7ba862-be40-467b-97af-da1eff4fc70e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== List all data variables in the Dataset ds2.\n",
+ "ds2.data_vars\n",
+ "\n",
+ "list(ds2.data_vars)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2ee0a683-b8e4-43b0-b172-0e08c0337642",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Look at the newly created Dataset ds2 in a \"ncdump\"-like fashion.\n",
+ "ds2.info()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "81bb750f-ddc1-4c78-a063-a3eae89a0c96",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Access the data variable temp.\n",
+ "myvar = \"temp\"\n",
+ "ds2[myvar].head(2)\n",
+ "# equivalent alternatives:\n",
+ "#ds2[\"temp\"].head(2)\n",
+ "#ds2.temp.head(2)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "554cb5a6-5b17-4cea-81e8-998d8e2f13fd",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Access the latitude coordinate variable associated with the temp variable in the xarray Dataset.\n",
+ "ds2.temp.lat\n",
+ "#== Alternative:\n",
+ "#ds2[\"temp\"].lat\n",
+ "#ds2[myvar].lat"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "70ee7dfb-fcf2-4c1c-85b7-57a13ee15f3f",
+ "metadata": {},
+ "source": [
+ "_**Note**_:In case of ds2, the latitude coordinate variable associated with the temp variable is identical \n",
+ "with the latitude coordinate variable associated with xarray Dataset.\n",
+ "Therefore, ds2.lat could simply be used instead."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "048e197e-4e8d-4928-87e0-0a4ac93f3b68",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Get specific information about the dimensions of a Dataset.\n",
+ "print(f\"dimensions: {ds2.dims}\") \n",
+ "#== Get specific information about the size and shape of the data variable temp in the Dataset.\n",
+ "print(f\"size: {ds2.temp.size}\")\n",
+ "print(f\"shape: {ds2.temp.shape}\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "2fe9f2dd-c701-4137-b1c1-bde828643ef2",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 10: Construct an xarray Dataset by merging two xarray DataArrays"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "472a4098-89fc-4ab9-94ee-51bf36aa1775",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Generate two xarray DataArrays.\n",
+ "\n",
+ "da5 = xr.DataArray(mydata, \n",
+ " name=\"var1\", \n",
+ " dims=[\"time\", \"lat\", \"lon\"], \n",
+ " coords=[tcoords, ycoords, xcoords]) #-- 3D DataArray\n",
+ "da6 = xr.DataArray(mydata[-1,1:-1,:], \n",
+ " name=\"var2\",\n",
+ " dims=[\"lat2\", \"lon\"], \n",
+ " coords=[ycoords[1:-1], xcoords],\n",
+ " attrs={\"units\":\"K\"}) #-- 2D DataArray"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "23f63ea0-40d4-4843-802c-0c0a4bf2d9c2",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Merge them with xr.merge(). This function call returns a xarray Dataset.\n",
+ "\n",
+ "ds3 = xr.merge([da5, da6])\n",
+ "#== NOTE: Merging only works if each DataArray has a name attribute!\n",
+ "# As alternative, you can merge DataArrays with the xr.Dataset function like, e.g.\n",
+ "# ds = xr.Dataset({\"airtemp\": da5, \"cantemp\": da6})"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "dbba060c-94c4-4c73-8de8-729a73928b09",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Add some global attributes. It is advisable to add a title, a history, institution, references, etc. \n",
+ "ds3.attrs[\"history\"] = \"myhistory\""
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f3f428e9-76be-455e-84e3-b2c71e81eced",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Print the new Dataset. var1 and var2 only share the dimensions lon. \n",
+ "ds3"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "1adb8c2a-4efa-42ab-a032-d32478fe9a70",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "_**NOTE**_: You can also merge a DataArray to an existing Dataset with `xr.merge()`."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6f2ac0ea-27c9-4544-81ae-230bae27ea36",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "ds4 = xr.merge([ds3, da6.rename(\"var3\")])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "d9bef20d-18f2-4692-8c9e-fe5b1d3e47a6",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== With the following command, you can list both data variables and coordinates names of the Dataset.\n",
+ "#== var3 is now also included\n",
+ "list(ds4.variables)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f2a449d6-05a3-4268-bb20-9112d69f0f1f",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "## _(C) Indexing and slicing with xarray data_\n",
+ "***"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "8ca2cef7-79dd-4a3e-b4f2-72d415dca6b4",
+ "metadata": {},
+ "source": [
+ "**Overview of the indexing options** <br>\n",
+ "see also: https://docs.xarray.dev/en/stable/user-guide/indexing.html#\n",
+ "\n",
+ "| Dimension lookup | Index lookup | DataArray syntax | Dataset syntax |\n",
+ "|:-----------------|:--------------|:-------------------------------------------------|:-------------------------------------------------|\n",
+ "| Positional | By integer | `da[0, :, :]` | not available |\n",
+ "| Positional | By label | `da.loc[\"2001-01-01\", :, :]` | not available |\n",
+ "| By name | By integer | `da.isel(time=0)` or <br> `da[dict(time=0)]` | `ds.isel(time=0)` or <br> `ds[dict(time=0)`] |\n",
+ "| By name | By label | `da.sel(time=\"2001-01-01\")` or <br> `da.loc[dict(time=\"2001-01-01\")`] | `ds.sel(time=\"2001-01-01\"`) or <br> `ds.loc[dict(time=\"2001-01-01\")]` |"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "d2bf2c7b-8e77-43be-8a1b-87c4497c90a2",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 11: Indexing and Slicing on an xarray DataArrays"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "382a6620-1213-43d2-9e21-e360338ac2cb",
+ "metadata": {},
+ "source": [
+ "For demonstration, we create an xarray DataArray of shape(4,6). <br>\n",
+ "The dimension **x** can be interpreted as **row** and the dimension **y** as **column**."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "dcbd2a1e-9d4f-4678-95fe-2189d3e736cb",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "da7 = xr.DataArray(np.arange(1,25).reshape((4,6)),\n",
+ " dims=[\"x\", \"y\"],\n",
+ " coords={\"x\":[1,2,3,4], \"y\":[10,20,30,40,50,\"A\"]})\n",
+ "da7"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "d6477116-6657-4776-9de4-1743a9dc9112",
+ "metadata": {},
+ "source": [
+ "#### (a) Positional, by integer\n",
+ "\n",
+ "Indexing can be done by the index(es) of the dimension(s).\n",
+ "\n",
+ "The general syntax for accessing values in a two-dimensional DataArray using two indices is:\n",
+ " `data_array [index_of_dim_0][index_of_dim_1]` \n",
+ "or \n",
+ " `data_array [index_of_dim_0, index_of_dim_1]`\n",
+ " \n",
+ "The general syntax for accessing values in a two-dimensional DataArray using a range of indices along one or more dimensions (_slicing_).<br>\n",
+ "`data_array[start_index_0:end_index_0][start_index_1:end_index_1]` \n",
+ "or \n",
+ "`data_array[start_index_0:end_index_0, start_index_1:end_index_1]` \n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "285652a6-8adf-477e-8ec0-5fcee1c3b65e",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Retrieve the value from the data array at the index positions [1][0], \n",
+ "#== corresponding to the second row and the first column in the data array da7.\n",
+ "da7[1][0]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "22f7b17d-8451-41b8-8ba9-bf28bd356282",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== When only a single index is provided for slicing, \n",
+ "#== it operates along the first dimension of the data array. \n",
+ "#== Using da7[0], the entire first row (i.e. the first dimension x) of da7 is selected.\n",
+ "da7[0]"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "ab86532c-f47b-4e1f-921d-26069ca84886",
+ "metadata": {},
+ "source": [
+ "#### (b) By name, by integer\n",
+ "\n",
+ "Indexing with the `.isel()` method uses the dimension name and the integer index.<br>\n",
+ "Slicing with the `.isel()` method uses the dimension name and the integer index range indicated with `slice`."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "98d4c335-da25-4641-a265-2c44b8e4b0a7",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Select the second row and the first column of the data array da7.\n",
+ "da7.isel(x=1, y=0)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f47628dc-b33d-4ec2-b18a-e1f049cba88b",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Slice the first and second row and the second and third column of da7.\n",
+ "da7.isel(x=slice(0,2), y=slice(1,3))\n",
+ "# This is equivalent to\n",
+ "# da7[0:2, 1:3]"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "cd013b19-fc63-4125-bec0-9a067a4355e6",
+ "metadata": {},
+ "source": [
+ "### (c) By label, by name\n",
+ "\n",
+ "With the `.sel()` method, you can select specific values from a DataArray based on _coordinate values_.<br>\n",
+ "This can also be done with the `.loc[]` method."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e59aad98-1af1-4e60-9648-da66ea4ea5e6",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Selecting a value from da7 at the coordinates x=3 and y=\"10\". \n",
+ "da7.sel(x=3, y=\"10\") # By default, sel requires exact matches.\n",
+ "#== Alternatively, you can use .loc[]\n",
+ "da7.loc[{\"y\": \"10\", \"x\": 3}]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "fc26f1be-c990-4c19-a321-4efcf02ee4d7",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Selecting a values from da7 at the coordinates closest to x=3.4\n",
+ "# using nearest neighbor\" interpolation (method=\"nearest\")\n",
+ "da7.sel(x=3.4, method=\"nearest\")\n",
+ "# you can also specify a maximum distance for inexact matches using tolerance.\n",
+ "# da7.sel(x=3.4, method=\"nearest\", tolerance=0.5)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "74de761c-ab70-442c-b336-fa61d268bf91",
+ "metadata": {},
+ "source": [
+ "_**Note**_: `.loc[]` does not offer specifying a method!"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f2e5cc5a-4f0e-4d9f-8629-55672db6411d",
+ "metadata": {},
+ "source": [
+ "### (d) Positional, by label\n",
+ "\n",
+ "With the `.loc[]` method, you can select specific values from a DataArray based on coordinate values. "
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "3bfd79a3-6b78-4412-8e8b-d8c5578ed375",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "da7.loc[1:3, \"A\"]"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "303ce799-6f25-4c6d-b78f-9b3f82c28242",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 12: Indexing and Slicing on an xarray Datasets"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "5f73845e-2463-49ca-9b1d-fc3590c742bf",
+ "metadata": {},
+ "source": [
+ "Indexing resp. slicing on xarray Datasets can only be done using a _by name_ dimension lookup in combination with an index lookup `by integer` or `by label`. "
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f631e9e9-2e90-4e12-8c75-61ed3e987551",
+ "metadata": {},
+ "source": [
+ "For demonstration, we use the xarray Dataset ds4 from Showcase 10. "
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "d4a0dd89-9de5-4840-a4b9-d348c0a3d8eb",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds4"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "03f27e2e-48c4-44da-b391-374efbfe8ace",
+ "metadata": {},
+ "source": [
+ "### (a) By name, by label"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "b8f52aab-1557-4d8a-ac37-7e61d4087f6a",
+ "metadata": {},
+ "source": [
+ "Using the .sel() method directly on the Dataset affects all contained data variables, applying the selection along the shared coordinates."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "5c9ad0ac-3342-45bb-a88e-4f7c0088d742",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Extract data for the time step 2022-03-02 from all variables in ds4\n",
+ "ds4.sel(time=\"2022-03-02\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2c2cbec2-118c-490a-976f-436cda185dfc",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Extract data from the variable var1 for the time step 2022-03-02\n",
+ "ds4.var1.sel(time=\"2022-03-02\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "4b1d2dd2-c408-4646-b3f1-bb91ed177c27",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds4.var1.sel(time=\"2022-03-02\",lon=slice(1,3)).values"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f85e1b34-3c38-4bdf-875c-6b97c492e40c",
+ "metadata": {},
+ "source": [
+ "Using the `.loc()` method, you can use a dictionary to provide the indexing/slicing argument"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e9da4f10-c1f7-469c-8534-4f79d18358d2",
+ "metadata": {},
+ "source": [
+ "If you would prefer to work more Pandas-like, then you can use the .loc[] method that uses a dictionary."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "68397391-7fc1-4563-a556-310f2f7ce59c",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds4.loc[{\"time\":\"2022-03-02\", \"lon\": slice(1, 4)}]"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "62498905-f033-4c0c-a28c-3ec1bd6f3b95",
+ "metadata": {},
+ "source": [
+ "<br>\n",
+ "\n",
+ "### Exercise 4: Indexing and Slicing"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "9c22ebd0-af37-4643-93a7-c7784d79d333",
+ "metadata": {},
+ "source": [
+ "Extract some data from the Dataset ds4 from Showcase 10 using<br>\n",
+ " 1. .isel()\n",
+ " 2. .sel()\n",
+ " 3. .loc[]\n",
+ "Apply these functions both on the entire Dataset and data variables \n",
+ "Which method do you like better `.sel()` or `.loc[]`?"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "3370f2ba-13c4-4336-afd8-07d06099b77c",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 1\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "56992a3b-c3ee-46c3-bd2d-ea6acae788ab",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 2\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "9b6f873b-6cb1-40c0-9322-7212d938310c",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 3\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "edcaf3ab-4bc3-4520-ab00-06b92cc3ead9",
+ "metadata": {},
+ "source": [
+ "#### Solution Exercise 4"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "c374864d-09fa-4c28-a124-e5f47a2f5f68",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 1 on the Dataset\n",
+ "ds4.isel(lat=2,lon=0,time=1).values"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "93112261-32ca-4907-a17b-1c1922aa42a1",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 1 on a data variable\n",
+ "ds4.var2.isel(lat2=2,lon=0).values"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "9818597a-f2b6-453f-88da-f1a9f752d0e3",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 2 on the Dataset\n",
+ "ds4.sel(lat2 = 31, time = \"2022-03-02\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e452778c-d4db-4435-9e98-a9b3342529e9",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 2 on the data variable\n",
+ "ds4.var3.sel(lat2 = slice(32,34.4))"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "2e8be05b-6657-4c4f-856d-07b7e2d1fb6b",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 3 on the Dataset\n",
+ "ds4.loc[{\"lat2\": [31, 34], \"time\": \"2022-03-02\"}]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "a3620a16-56b5-4754-b081-daddf46d8675",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 3 on the data variable\n",
+ "ds4.var3.loc[{\"lat2\": slice(31, 34)}]"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "bdc2e17f-0636-4e7a-a4b1-b73a8661a257",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "## _(D) Save, open, and read files with xarray_\n",
+ "\n",
+ "***\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "566e78fe-7c7d-41c1-a645-74a9aefe15ce",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 13: Saving xarray DataArrays or Datasets as netCDF file"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "b68c908b-b3ec-45d2-9bfd-04b08d50b677",
+ "metadata": {},
+ "source": [
+ "\n",
+ "The `to_netcdf` method allows you to save a Dataset or DataArray to a netCDF file.<br> The default mode is \"w\" (write), but you can specify other modes such as \"a\" (append) or \"r\" (read) as arguments to the function call. "
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "fa43249d-bf4e-4196-8081-2d78f64a4d6c",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Save the xarray Dataset ds4 from Showcase 10.\n",
+ "ds4.to_netcdf(\"ds4.nc\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "719e1bdc-33d9-4e7d-b18f-997efb317472",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "!ncdump -h ds4.nc"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "0d6f0c0c-e08f-49c9-90cd-996022f4a797",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Save the xarray DataArray da6 from Showcase 10.\n",
+ "da6.to_netcdf(\"da6.nc\", mode=\"a\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "94c5b03e-b362-495c-b543-2787a8cd06a8",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "!ncdump -h da6.nc"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "aaaf5cba-b948-4e3e-8c99-cb975d88a71e",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 14: Open and read a netCDF file with xarray"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "a0188112-5f2f-49a0-9bcc-46919e05c49d",
+ "metadata": {},
+ "source": [
+ "xarray provides the function `xr.open_dataset()` to open a file with the file format netCDF, GRIB, HDF5, or Zarr. Default format is netCDF.<br>\n",
+ " `ds_in = xr.open_dataset(\"infile.nc\")`<br>\n",
+ "is the same as<br>\n",
+ " `ds_in = xr.open_dataset(\"infile.nc\", engine=\"netCDF4\")`"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "61db211a-a491-45c7-a55b-6ac77386d5e0",
+ "metadata": {},
+ "source": [
+ "<div class=\"alert alert-info\">\n",
+ " <b>**xarray is Lazy**:</b> By default, xarray performs lazy loading: With open_dataset, it only loads the metadata (such as variable names, dimensions, and attributes) from the netCDF file into memory, without loading the actual data values.<br>\n",
+ "The data values are loaded into memory only when you explicitly access them or perform operations that require accessing the data. Lazy loading helps saving memory and enhancing the performance.\n",
+ "</div>\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "2bc976b1-809d-4615-af4f-c3e600325119",
+ "metadata": {},
+ "source": [
+ "## Open and read a netCDF file"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "7f2dc515-49b3-4cdc-89fe-81131bdf520b",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Open (lazy-loading) the netCDF file created in Showcase 13.\n",
+ "xr.open_dataset(\"ds4.nc\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "15f07932-b524-4212-b8b0-4853f13f1709",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Open the netCDF file created in Showcase 13 and load the data into memory.\n",
+ "ds5 = xr.open_dataset(\"ds4.nc\").load()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "87065f99-cffd-40b5-8831-7fad9686c293",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "ds5"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "1c3ba127-ba60-4b15-bd73-5e1b4d88f791",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== In case you wish to remove the reference to the Dataset ds5, use:\n",
+ "# del(ds5)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "1ad5e56e-190c-44b2-af52-2243f7b4c4b2",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Open ds4.nc and then lazily loading only the variables \"var1\" and \"var2\" from it.\n",
+ "ds6 = xr.open_dataset(\"ds4.nc\")[[\"var1\", \"var2\"]]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "04cfda1e-d54d-49c1-b89b-70ff32c75038",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds6"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "2424d12e-3433-42f2-9d39-9edd24d23290",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 15: Open and read a GRIB file with xarray\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "c496f4c5-f38e-44a9-80f1-ffc8ba003fce",
+ "metadata": {},
+ "source": [
+ "By default, the xr.open_dataset() function selects the _engine_ (_i.e. library or backend used to read the file_) to open the file based on the file name extension or attempts to infer it the file format based on the file contents. <br>\n",
+ "For GRIB files, the engine is usually `cfgrib`."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "b4a0b3a0-7cfa-4651-889b-4f00a283af94",
+ "metadata": {},
+ "source": [
+ "_**NOTE**_: when opening a GRIB file with cfgrib as the engine in xarray, an index file (.idx) is typically created or used if available. If an available index file is incompatible with the GRIB file, xarray issues a warning indicating that the index file is being ignored. "
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "81ffa90e-d4b8-46e9-a361-5a08bfa51f99",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "ds7 = xr.open_dataset(\"../data/MET9_IR108_cosmode_0909210000.grb2\")\n",
+ "# same as \n",
+ "# ds7 = xr.open_dataset(\"../data/MET9_IR108_cosmode_0909210000.grb2\", engine=\"cfgrib\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "d11b846a-c077-4afa-ac08-8208bf8d9cfc",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#-- List the data variables\n",
+ "print(f\"{ds7.data_vars} \\n {ds7.coords}\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "69443de1-d3cd-4f80-a76e-9a392dd73d11",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 16: Open and read a multiple netCDF files at once with xarray\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "43ffefd1-b40b-4c9e-8901-3f127d1b2e9e",
+ "metadata": {},
+ "source": [
+ "*xarray* provides the function `xr.open_mfdataset()` to read multiple files in one step as a single Dataset. \n",
+ "When using `xr.open_mfdataset()`, it recommended to have **dask** enabled (by having dask installed in your environment). **Dask** is utilized to load and process the data in a parallelized manner. "
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f5559513-bb2b-43d6-9054-4438261d2619",
+ "metadata": {},
+ "source": [
+ "For demonstration, we open three netCDF files _precip_day01.nc, precip_day02.nc, and precip_day03.nc_, each containing the data of one day in 6 hour intervals. "
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "1cc4ab67-5444-4c1b-bfe7-6733e94a96f8",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Open the files with xr.open_mfdataset\n",
+ "ds8 = xr.open_mfdataset(\"../data/precip_day*.nc\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "d3cc7d3b-b518-4c4c-90c1-8dae6025d314",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "_**NOTE**_: By default, xr.open_mfdataset will concatenate the dimensions of the Datasets along which they overlap.\n",
+ "In this example, the data are concatenated along the time dimension.\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "8592569c-470c-4fa2-bbc1-428c30c1c366",
+ "metadata": {},
+ "source": [
+ "<div class=\"alert alert-info\">\n",
+ " <b>Dask is Very Lazy!</b> <br>\n",
+ " When Dask is used to load the data, the data are loaded as dask.array objects.\n",
+ "</div>"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "7ef53a23-a737-48a8-aaa3-f73467276e4e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Look at the description of the data variable of ds8. \n",
+ "#== You will see that precip is represented as dask.array object.\n",
+ "ds8"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f52cfbbb-e6f1-4beb-81f8-c5df80e9cbc4",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Indexing dask.array objects in ds8 will not directly display the exact values, \n",
+ "#== but instead provide a preview of the data.\n",
+ "#== To access a specific point in the array, you need to load the data into memory first.\n",
+ "ds8.precip[1,4,5]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "be82b529-91ec-4106-ab5f-296fae96a563",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Loading the accessed data point as xarray DataArray. \n",
+ "ds8.precip[1,4,5].load()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "0d740310-1c01-4360-a10e-e07287c7153e",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Alternative: loading the accessed data point as NumPy array.\n",
+ "ds8.precip[1,4,5].values"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "74da3f4d-efa8-404a-8835-a34acb65ac04",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== You can also load the entire Dataset or individual data variables.\n",
+ "ds8.load().head(2)\n",
+ "# or\n",
+ "#ds8.precip.load().head(2)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e3e2cb78-16a5-408e-9972-009a24d6728e",
+ "metadata": {},
+ "source": [
+ "<br>\n",
+ "\n",
+ "***\n",
+ "\n",
+ "### Exercise 5: Open a netCDF file with `xr.DataArray`\n",
+ "\n",
+ "<br>"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "300aba36-5fea-4a1e-bf0a-cbb794cbe21e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 1. Open the file \"../data/rectilinear_grid_2D.nc\" with xarray\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "cfb0bb07-a02a-4dbf-8b90-993bba39b55b",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 2. List all data variables of the Dataset as a list\n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "83afc2ad-daf3-485b-bcc4-75ecf430eba0",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 3. List all data variables and coordinate variables of the Dataset as a list\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "d935b15e-23f0-4bbe-ac76-16d83c65eaee",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 4. Lazy load the first variable of the list, print it with head(2) \n",
+ "# Then load it into memory while assigning it to a variable da_rg. \n",
+ "# Print it again with head(2) \n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "429b43e4-2321-4cf0-bc58-48bd459468f3",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# 5. Plot the first timestep of da_rg\n",
+ "\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "60b1ce25-8fcb-49b4-a3a1-758b174363ad",
+ "metadata": {
+ "tags": []
+ },
+ "source": [
+ "#### Solution Exercise 5"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e87e1214-104e-42ae-9d1d-98808a2e2270",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 1.\n",
+ "ds9 = xr.open_dataset(\"../data/rectilinear_grid_2D.nc\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "8e257ce3-11b0-4384-be47-e53a9cd423d0",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 2.\n",
+ "list(ds9.data_vars)\n",
+ "list(ds9.keys())\n",
+ "list(ds9)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "5890c3f2-9810-4657-a441-640efbd9c656",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 3.\n",
+ "list(ds9.variables)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "9d657253-7029-48d8-a661-1b36bcc1fe4a",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 4.\n",
+ "ds9[list(ds9)[0]].head(2)\n",
+ "print(\"*\"*100)\n",
+ "da_rg = ds9[list(ds9)[0]].load()\n",
+ "da_rg.head(2)\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f2b329a4-2bad-4960-8659-2377e99e6651",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "# 5.\n",
+ "da_rg.isel(time=0).plot();"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "10b0181a-9436-40fb-b771-9ff8ff8d9773",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "## _(E) Appendix_\n",
+ "***"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "61d518b4-7410-46f3-9480-cee9bb1e06e5",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 17: Expand the dimensions of a xarray DataArray\n"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f109a858-3383-405f-8794-eaf160745328",
+ "metadata": {},
+ "source": [
+ "With `DataArray.expand_dims()`, you can add a new dimension, e.g. time, to a DataArray A. This function call returns a new DataArray object. \n",
+ "In the new, expanded DataArray B, the original values of A are repeated along the new dimension (e.g. time). In other words, the original values of A are broadcasted along this new dimension to match the shape of the resulting DataArray B."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "f1e51bca-4e86-42dc-9915-89054f10e6ec",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Add a time dimension with values 1 and 2 to the pwr_da5 DataArray.\n",
+ "time = [1, 2]\n",
+ "pwr_da6 = pwr_da5.expand_dims({\"time\":time}, axis=0) \n",
+ "# Note: axis=0 ensures that the new dimension is inserted as the first dimension, \n",
+ "# so that the resulting dimension order is time,lat,lon."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "50af7478-0672-48b3-a5a8-6df1f8202aa8",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Compare the dimensionality and shape of the old and new DataArray:\n",
+ "\n",
+ "print(f\" dims before: {pwr_da5.dims}\\n dims after: {pwr_da6.dims}\")\n",
+ "print(\"====\"*20)\n",
+ "print(f\" shape before: {pwr_da5.shape}\\n shape after: {pwr_da6.shape}\")\n",
+ "print(\"====\"*20)\n",
+ "print(f\" sizes before: {pwr_da5.sizes}\\n sizes after: {pwr_da6.sizes}\")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6da14987-2dcb-4291-bb57-efa2e88e7c7e",
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
+ "source": [
+ "#== Compare if the data values at the first time step are equal to the second time step. \n",
+ "\n",
+ "import numpy.testing as npt\n",
+ "try:\n",
+ " npt.assert_array_equal(pwr_da6.isel(time=0).data, pwr_da6.isel(time=1).data)\n",
+ " print(\"Equal\")\n",
+ "except AssertionError:\n",
+ " print(\"Not equal\")"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "3d546977-6354-4fe6-bf3f-6c0560895554",
+ "metadata": {},
+ "source": [
+ "***\n",
+ "\n",
+ "### Showcase 18: Add a new coordinate variable to an xarray DataArray\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "131bdc62-72fa-4855-9672-5c0d0e2c6168",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== Add a new coordinate variable named \"station_name\" in the prw_da2 DataArray. \n",
+ "#== This new coordinate variable contains the station names provided in the prw_stations array.\n",
+ "prw_da2[\"station_name\"] = xr.DataArray(prw_stations, dims=\"Station\")\n",
+ "prw_da2.head(5)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "cdf8fc95-481d-479d-a3a5-07391939961f",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#== When plotting, we can now use the new coordinate variable to label the x-axis.\n",
+ "prw_da2.plot(x=\"station_name\", figsize=(16,4));\n",
+ "plt.xticks(rotation=90);\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e9fe3bc1-3a11-400f-ad52-cf70230e40f8",
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "0 Python 3 (based on the module python3/unstable",
+ "language": "python",
+ "name": "python3_unstable"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.10.10"
+ },
+ "toc-autonumbering": false
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/notebooks/xarray_introduction_part2.ipynb b/notebooks/xarray_intro_part2.ipynb
similarity index 62%
rename from notebooks/xarray_introduction_part2.ipynb
rename to notebooks/xarray_intro_part2.ipynb
index 6b9cbbe1bf1e0f97c422cc5e48b2baea6de13250..e6c748e981e4192e34d0437d5e588ad5b0e9f1d7 100644
--- a/notebooks/xarray_introduction_part2.ipynb
+++ b/notebooks/xarray_intro_part2.ipynb
@@ -23,62 +23,50 @@
"source": [
"\n",
"<p align=\"center\">\n",
- " <img src=\"https://docs.xarray.dev/en/stable/_static/Xarray_Logo_RGB_Final.svg\" width=\"35%\" align=\"right\" title=\"Xarraylogo\" hspace=\"20\">\n",
+ " <img src=\"https://docs.xarray.dev/en/stable/_static/Xarray_Logo_RGB_Final.svg\" width=\"35%\" align=\"right\" title=\"xarraylogo\" hspace=\"20\">\n",
"</p>\n",
"\n",
- "<font size=\"20\"> Xarray Introduction II</font> \n",
+ "<font size=\"20\"> xarray introduction II</font> \n",
"\n",
- "Xarray home page: https://xarray.pydata.org/en/stable/index.html <br>\n",
- "Xarray documentation: https://docs.xarray.dev/en/stable/index.html\n"
+ "xarray documentation: https://docs.xarray.dev/en/stable/index.html\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "# How to create a Xarray DataSet with two 3D data variables "
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {},
- "source": [
- "## Importing required libraries"
+ "<br>\n",
+ "\n",
+ "========================================================================================\n",
+ "# Importing Modules and Configure the Notebook\n",
+ "========================================================================================"
]
},
{
"cell_type": "code",
"execution_count": null,
- "metadata": {},
+ "metadata": {
+ "tags": []
+ },
"outputs": [],
"source": [
"import xarray as xr\n",
"import numpy as np\n",
"import pandas as pd\n",
- "import datetime\n",
"import matplotlib.pyplot as plt\n",
- "import os, datetime"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
+ "import matplotlib as mpl\n",
+ "import os, datetime\n",
+ "\n",
+ "\n",
+ "# Set this to render all evaluated output of a cell\n",
"from IPython.core.interactiveshell import InteractiveShell\n",
- "InteractiveShell.ast_node_interactivity = \"all\""
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {
- "tags": []
- },
- "source": [
- "### Define your fake data"
+ "InteractiveShell.ast_node_interactivity = \"all\"\n",
+ "\n",
+ "# Set default figure size and font size\n",
+ "mpl.rcParams.update({\n",
+ " \"figure.figsize\": (3.5, 2.5),\n",
+ " \"font.size\": 9\n",
+ "})"
]
},
{
@@ -87,45 +75,31 @@
"tags": []
},
"source": [
- "as lists for the coordinate variables of time, lat, lon"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "time = ['2023-01-01', '2023-01-02']\n",
- "lat = [45.,50.,55.,60.]\n",
- "lon = [0.,5.,10.,15.,20.]"
+ "<br>\n",
+ "\n",
+ "======================================================================\n",
+ "# Showcases and Exercises\n",
+ "======================================================================\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "create some random 3D numpy ndarray representing your dummy data"
+ "***\n",
+ "## _(A) xarray functions/methods: where()_\n",
+ "***\n"
]
},
{
- "cell_type": "code",
- "execution_count": null,
+ "cell_type": "markdown",
"metadata": {
"tags": []
},
- "outputs": [],
"source": [
- "mydata1 = np.random.uniform(250,300,40).reshape((len(time),len(lat),len(lon)))"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {},
- "source": [
- "check the shape of your dummy data"
+ "***\n",
+ "\n",
+ "### Showcase 1: Create a xarray Dataset by open a netCDF file"
]
},
{
@@ -136,24 +110,9 @@
},
"outputs": [],
"source": [
- "mydata1.shape"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {
- "tags": []
- },
- "source": [
- "### Create the first xarray DataArray (i.e. the first variable)"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {},
- "source": [
- "This is done by converting the ndarray dummy data plus the coordinate variables into an xarray DataArray. \n",
- "Coordinates are added as dictionary in a key:value manner"
+ "#== Open the the file rectilinear_grid_2D.nc (see also Exercise 5 part 1) with xr.open_dataset().\n",
+ "#== Select only the first time step and the data variables tsurf and precip \n",
+ "ds1 = xr.open_dataset(\"../data/rectilinear_grid_2D.nc\").isel(time=0)[[\"tsurf\", \"precip\"]]"
]
},
{
@@ -164,19 +123,8 @@
},
"outputs": [],
"source": [
- "da1 = xr.DataArray(data=mydata1,\n",
- " coords={'time': time,\n",
- " 'lat': lat,\n",
- " 'lon': lon,\n",
- " },\n",
- " attrs={'units': 'K', 'standard_name':'air_temperature'})"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {},
- "source": [
- "Check the DataArray by printing"
+ "#== Inspect the Dataset\n",
+ "ds1"
]
},
{
@@ -187,77 +135,41 @@
},
"outputs": [],
"source": [
- "da1"
+ "#== plot some data\n",
+ "ds1.tsurf.isel(lat=slice(1,4)).plot.line(x=\"lon\",marker='*',markersize=2);"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "check the first time step of the data by plotting"
+ "***\n",
+ "\n",
+ "### Showcase 2: Masking data with `where()`"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "For 2D data, the default figure type of the plot() method is a pcolormesh. \n",
- "In this case, the output of plot() and plot.pcolormesh() is identical. \n",
- "see https://docs.xarray.dev/en/stable/generated/xarray.DataArray.plot.html"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "da1.isel(time=0).plot(aspect=2, size=2); \n",
+ "Xarray provides a `where()` function that uses one or several conditions to filter data. <br>\n",
+ "With `where()`, you can e.g. \n",
+ "* filter the data by data value range<br>\n",
+ "or <br>\n",
+ "* by a condition related to a dimension.\n",
+ "<br>\n",
"\n",
- "#-- the plot() method offers many configurations options of the plot, e.g. cmap=\"Blues\"\n",
- "#da1.isel(time=0,lat=slice(1,4)).plot(cmap=\"Blues\")"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# you can also plot a histogram\n",
- "da1.isel(time=0).plot.hist(aspect=2, size=2);\n",
- "plt.title('mytitle');"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {
- "tags": []
- },
- "source": [
- "you can also plot 2D data as a line plot with the plot.line() method, provided you specifiy one dimension as either x or y, which are then used as an axis of the line plot. \n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "da1.isel(time=0,lat=slice(1,4)).plot.line(x=\"lat\",marker='*',markersize=10, aspect=2, size=2);"
+ "The **`where()`** function returns a new DataArray where values that \n",
+ "* **satisfy the condition are preserved**, <br>\n",
+ "and\n",
+ "* do not satisfy the condition are replaced with NaN by default."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "### Create the second xarray DataArray (i.e. the second variable)"
+ "**Filter data by data range using `where()` in a single condition**"
]
},
{
@@ -266,8 +178,9 @@
"metadata": {},
"outputs": [],
"source": [
- "#-- create a new DataArray by inheriting the attributes, dims, coords from da1\n",
- "da2 = da1.copy(data=np.cos(mydata1)+10)"
+ "#== Select DataArray to be used for where()\n",
+ "myvar = \"tsurf\"\n",
+ "var = ds1[myvar]"
]
},
{
@@ -276,34 +189,28 @@
"metadata": {},
"outputs": [],
"source": [
- "#-- overwrite the standard name inherited from da1\n",
- "da2.attrs[\"standard_name\"] = \"canopy_temperature\""
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {},
- "source": [
- "### Convert the DataArrays into a DataSet"
+ "#== Data values not meeting the condition are set to nan.\n",
+ "da1_extremes = var.where(var > 296.) "
]
},
{
"cell_type": "code",
"execution_count": null,
- "metadata": {
- "tags": []
- },
+ "metadata": {},
"outputs": [],
"source": [
- "# it is mandatory to specify a name for each DataArray\n",
- "ds = xr.Dataset({'airtemp': da1, 'cantemp': da2})"
+ "#== Inspect the masked data.\n",
+ "da1_extremes.values[:2,:2]\n",
+ "var.shape, da1_extremes.shape"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "### Look at the DataSet"
+ "--> The masked data have the same shape as the original data.<br> \n",
+ "--> The masked values are represented as nan, the unmasked values remain unaltered.<br> \n",
+ "--> The masked data array is of type float32 (da1_extremes.dtype): <br>"
]
},
{
@@ -314,21 +221,24 @@
},
"outputs": [],
"source": [
- "ds"
+ "da1_extremes.plot();"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "### Edit the global metadata"
+ "***\n",
+ "\n",
+ "### Showcase 3: Masking data with Python boolean expression and `where()`"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "this is the metadata describing the entire dataset. For example, add the CF conventions, if you want to write out CF-compliant netCDF files in the end. In addition, it is advisable to add a title, a history, institution, references, etc. See also https://cfconventions.org/Data/cf-conventions/cf-conventions-1.10/cf-conventions.html section 2.6.2."
+ "A **boolean expression** is a common way to create **boolean masks** in Python. \n",
+ "These masks are essential for masking other data using the where() function."
]
},
{
@@ -339,9 +249,8 @@
},
"outputs": [],
"source": [
- "ds.attrs={\"Conventions\": \"CF-1.10\", \n",
- " \"title\": \"this is just fake data for python course\",\n",
- " \"institution\": \"German Climate Computing Center (DKRZ)\"}"
+ "#== Create a boolean mask where all values greater 296K are true and the others false.\n",
+ "da2_extremes = (var > 296.) "
]
},
{
@@ -352,7 +261,9 @@
},
"outputs": [],
"source": [
- "ds"
+ "#== Look at da2_extremes\n",
+ "da2_extremes.shape\n",
+ "da2_extremes.data.dtype"
]
},
{
@@ -363,28 +274,9 @@
},
"outputs": [],
"source": [
- "#-- you can also edit the metadata afterwards\n",
- "ds.lat.attrs={\"standard_name\": \"latitude\", \"units\": \"degrees_north\"} "
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {},
- "source": [
- "----\n",
- "\n",
- "# Xarray functions/methods\n"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "## `where` - to mask data\n",
- "\n",
- "Similar to the NumPy `where` function, Xarray provides a `where` function that uses a condition to filter data. You can, e.g. filter the data by data value range or by a condition related to a dimension.\n"
+ "#== Mask the ds1.precip data with the da2_extremes mask\n",
+ "da3_prc = ds1.precip\n",
+ "da3_prc_masked = da3_prc.where(da2_extremes)"
]
},
{
@@ -395,23 +287,17 @@
},
"outputs": [],
"source": [
- "# For simplicity, we extract the data of interest from the Dataset prior the masking"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {},
- "outputs": [],
- "source": [
- "var = ds[\"airtemp\"].sel(time=\"2023-01-02\")"
+ "#== Plot the first time step of da2_prc_masked\n",
+ "da3_prc_masked.plot();"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "### Filter data by data range using `where` in a combined condition"
+ "***\n",
+ "\n",
+ "### Showcase 4: Filter by a condition related to a dimension using `where()`"
]
},
{
@@ -420,89 +306,109 @@
"metadata": {},
"outputs": [],
"source": [
- "mask1 = var.where((var > 273.15) & (var < 300.))\n",
- "mask1"
+ "#== Mask all data values in 'var' where longitudes are less than 90° East.\n",
+ "da4_dim_f = var.where(var.lon > 90.) #-- original shape preserved with default drop=False\n",
+ "da4_dim_f.plot(); "
]
},
{
"cell_type": "code",
"execution_count": null,
- "metadata": {
- "tags": []
- },
+ "metadata": {},
"outputs": [],
"source": [
- "mask1.dtype"
+ "#== Mask and drop all masked data values in 'var' where longitudes are less than 90° East.\n",
+ "da4_dim_f2 = var.where(var.lon > 90., drop=True) \n",
+ "da4_dim_f2.plot();"
]
},
{
"cell_type": "markdown",
- "metadata": {},
- "source": [
- "--> the masked data have the same shape as the original data \n",
- "--> The masked data are of type float4 (mask1.dtype). The masked values are represented as nan. "
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
"metadata": {
"tags": []
},
- "outputs": [],
"source": [
- "mask1.plot(aspect=2, size=2);"
+ "***\n",
+ "## _(B) xarray functions/methods: isnull(), notnull(), fillna(), count()_\n",
+ "***\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "### The alternative masking to `where` "
+ "***\n",
+ "\n",
+ "### Showcase 5: Create a mask with `isnull()` and `notnull()`\n",
+ "\n",
+ "<br>"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "For using one mask as another mask, you need a bolean mask"
+ "The `isnull()` function can identify missing or NaN (Not a Number) values in a dataset. <br>\n",
+ "It returns a **boolean mask** with \n",
+ "* \"True\" for missing values\n",
+ "and \n",
+ "* \"False\" for non missing values. \n",
+ "Conversely, the `notnull() function` returns True for non-missing values and False otherwise."
]
},
{
"cell_type": "code",
"execution_count": null,
- "metadata": {
- "tags": []
- },
+ "metadata": {},
"outputs": [],
"source": [
- "mask2 = (var >= 280)\n",
- "mask2"
+ "#== Create a boolean mask from masked precipitation data da3_prc_masked from Showcase 4\n",
+ "#== Missing values are returned as TRUE\n",
+ "da5_prc_bool1 = da3_prc_masked.isnull()\n",
+ "#== Missing values are returned as FALSE\n",
+ "da5_prc_bool2 = da3_prc_masked.notnull()"
]
},
{
- "cell_type": "markdown",
+ "cell_type": "code",
+ "execution_count": null,
"metadata": {},
+ "outputs": [],
"source": [
- "#### Example: Mask out ds['cantemp'] with mask2 and plot"
+ "fig, axs = plt.subplots(1,2, figsize=(9, 2))\n",
+ "da5_prc_bool1.plot(ax=axs[0])\n",
+ "da5_prc_bool2.plot(ax=axs[1]);"
]
},
{
"cell_type": "code",
"execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#==Print the dtype of da5_prc_bool1\n",
+ "da5_prc_bool1.dtype"
+ ]
+ },
+ {
+ "cell_type": "markdown",
"metadata": {
"tags": []
},
- "outputs": [],
"source": [
- "ds['cantemp'].sel(time=\"2023-01-02\").where(mask2, drop=True).plot(aspect=2, size=2)"
+ "***\n",
+ "\n",
+ "### Showcase 6: Count non-missing with `count()`\n",
+ "\n",
+ "<br>"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "### Filter by a condition related to a dimension"
+ "The `count()` function is used to count the number of non-missing values along one or more dimensions.<br>\n",
+ "If no dimensions are specified, it counts non-missing values across all dimensions."
]
},
{
@@ -511,30 +417,20 @@
"metadata": {},
"outputs": [],
"source": [
- "vartmp= var.where(var.lon > 10.)\n",
- "#vartmp.plot();"
- ]
- },
- {
- "cell_type": "markdown",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "## `isnull` - check where missing values exist\n",
- "\n",
- "It returns a mask of True/False elements. \n",
- "\n",
- "Our _var_ variable does not contain missing values. We now define values below 273.15 as missing.\n"
+ "#== count the number of non-missing values in da3_prc_masked (Showcase 3)\n",
+ "da3_prc_masked.count().values"
]
},
{
"cell_type": "code",
"execution_count": null,
- "metadata": {},
+ "metadata": {
+ "tags": []
+ },
"outputs": [],
"source": [
- "var = var.where(var < 273.15)"
+ "#== zonal pattern of non-missing values: non-missing value count along the latitude\n",
+ "da3_prc_masked.count(dim='lat').values[76:116] # slice values in the tropics"
]
},
{
@@ -543,76 +439,70 @@
"metadata": {},
"outputs": [],
"source": [
- "#-- to count the missings: count()\n",
- "print(var.isnull().count().values) "
+ "#==Count the total number of non-missing values. True values are counted as 1, and False values are counted as 0.\n",
+ "print(f\"N non-missing: {da3_prc_masked.count().values}\")\n",
+ "print(f\"N missing: {da3_prc_masked.isnull().sum().values}\")\n",
+ "print(f\"N total values: {da3_prc_masked.size}\")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "<br>\n",
- "\n",
- "## `count` - count valid values\n",
+ "***\n",
"\n",
+ "### Showcase 7: Replace missing values with a constant with `fillna()`\n",
"\n",
- "Count the data that are not missing values."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "metadata": {},
- "outputs": [],
- "source": [
- "var.count().values #-- with .values, the output will be printed as xarray DataArray "
+ "<br>"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "## `notnull` - check where valid (\"non-missing\") values\n",
- "\n",
- "To check where valid (\"non-missing\") values are present, use `notnull`.\n"
+ "The `fillna()` function is used to replace missing values in a dataset with specified values. "
]
},
{
"cell_type": "code",
"execution_count": null,
- "metadata": {},
+ "metadata": {
+ "tags": []
+ },
"outputs": [],
"source": [
- "var.notnull().values"
+ "#== replace all missing values in the da4_dim_f (from Showcase 4) with the value -200.\n",
+ "da4_dim_f.fillna(-200).plot();"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "<br>\n",
- "\n",
- "## `fillna` - change missing value to a constant number"
+ "***\n",
+ "## _(C) xarray functions/methods: statistics_\n",
+ "***\n"
]
},
{
- "cell_type": "code",
- "execution_count": null,
- "metadata": {},
- "outputs": [],
+ "cell_type": "markdown",
+ "metadata": {
+ "tags": []
+ },
"source": [
- "var.fillna(0.01).plot(aspect=2, size=2);"
+ "***\n",
+ "\n",
+ "### Showcase 8: Working with statistical functions\n",
+ "\n",
+ "<br>"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "<br>\n",
- "\n",
- "## `min(), max(), mean(), sum(), std(), corr(), ...`\n",
"\n",
- "Xarray provides a lot of computational functions.\n"
+ "Xarray provides a lot of computational functions: `min(), max(), mean(), sum(), std(), corr(), ...`\n"
]
},
{
@@ -621,16 +511,19 @@
"metadata": {},
"outputs": [],
"source": [
- "print(f'min = {ds.airtemp.min().values:6.2f}, max = {ds.airtemp.max().values:6.2f}')"
+ "print(f\"mean = {var.mean().values:6.2f}\")\n",
+ "print(f\"std = {var.std().values:6.2f}\")"
]
},
{
"cell_type": "code",
"execution_count": null,
- "metadata": {},
+ "metadata": {
+ "tags": []
+ },
"outputs": [],
"source": [
- "print(f'std = {ds.airtemp.std().values:6.2f}')"
+ "print(f\"min= {var.min().values:.1f}, \\nmax= {var.max().values:.1f}, \\npercentiles= {var.quantile([0.1,0.25,0.5,0.75,0.90]).values.round(1)}\")"
]
},
{
@@ -639,15 +532,9 @@
"source": [
"<br>\n",
"\n",
- "# Exercise 1\n",
- "\n",
- "Use the precip data from the ../data/rectilinear_grid_2D.nc file:\n",
+ "### Exercise 1: Masking and xarray statistical functions\n",
"\n",
- "1. compute the mean of the variable precip over 'time' and plot it\n",
- "1. plot only the precip data > 0.0001\n",
- "1. count how many non-missing values exist after masking values <=0.0001)\n",
- "1. for the variable precip, compute the field mean, i.e. the mean over ('lat','lon') \n",
- "1. plot the field mean as timeseries line plot and add a centered title\n"
+ "Use the precip data from the ../data/rectilinear_grid_2D.nc file:\n"
]
},
{
@@ -656,7 +543,8 @@
"metadata": {},
"outputs": [],
"source": [
- "# 1."
+ "# 1. compute the mean of the variable precip over 'time' and plot it\n",
+ "\n"
]
},
{
@@ -665,7 +553,8 @@
"metadata": {},
"outputs": [],
"source": [
- "# 2."
+ "# 2. plot only the precip data > 0.0001\n",
+ "\n"
]
},
{
@@ -674,7 +563,8 @@
"metadata": {},
"outputs": [],
"source": [
- "# 3."
+ "# 3. count how many non-missing values exist after masking values <=0.0001)\n",
+ "\n"
]
},
{
@@ -685,7 +575,8 @@
},
"outputs": [],
"source": [
- "# 4."
+ "# 4. for the variable precip, compute the field mean, i.e. the mean over ('lat','lon') \n",
+ "\n"
]
},
{
@@ -696,7 +587,8 @@
},
"outputs": [],
"source": [
- "# 5."
+ "# 5. plot the field mean as timeseries line plot and add a centered title\n",
+ "\n"
]
},
{
@@ -705,7 +597,7 @@
"source": [
"<br>\n",
"\n",
- "## Solution Exercise 1\n"
+ "#### Solution Exercise 1\n"
]
},
{
@@ -725,7 +617,8 @@
"metadata": {},
"outputs": [],
"source": [
- "ds2m = ds2.precip.mean('time')"
+ "myvar = \"precip\"\n",
+ "ds2m = ds2[myvar].mean('time')"
]
},
{
@@ -746,7 +639,7 @@
"source": [
"# 2.\n",
"\n",
- "ds2m.where(ds2m > 0.0001).plot(); #-- to subset region: xlim=50, ylim=0"
+ "ds2m.where(ds2m > 0.0001).plot(ylim=(-10,20), xlim=(90,140)) # to ylim and ylim to subset region"
]
},
{
@@ -755,8 +648,9 @@
"metadata": {},
"outputs": [],
"source": [
- "# 3.\n",
+ "# 3. count non-missing values\n",
"\n",
+ "ds2m.count().values\n",
"print(ds2m.where(ds2m > 0.0001).count().values)"
]
},
@@ -766,7 +660,7 @@
"metadata": {},
"outputs": [],
"source": [
- "# 4.\n",
+ "# 4. the mean over ('lat','lon') \n",
"\n",
"ds2fm = ds2.precip.mean(('lat','lon'))"
]
@@ -778,21 +672,35 @@
"outputs": [],
"source": [
"# 5.\n",
- "\n",
- "ax = plt.axes()\n",
- "ds2fm.plot(ax=ax);\n",
- "ax.set_title('field mean', loc='center', fontsize = 20);"
+ "ds2fm.plot();\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "----\n",
+ "***\n",
+ "## _(D) Advanced xarray: Climatology_\n",
+ "***"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "***\n",
"\n",
- "# Advanced Xarray Example: Climatology \n",
+ "### Showcase 9: \n",
"\n",
- "For our next example, we use a historical and scenario global time series from CMIP6."
+ "<br>"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "For our next example, we use a historical and scenario global time series from the climate model simulations <br>\n",
+ "conducted within CMIP6."
]
},
{
@@ -837,8 +745,52 @@
"metadata": {},
"outputs": [],
"source": [
- "dsh = xr.open_dataset(dir_data + fnameh)\n",
- "dsh"
+ "dsh = xr.open_dataset(dir_data + fnameh)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "metadata": {},
+ "source": [
+ "Check the data"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "#dsh.head(3)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "dsh.data_vars"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "dsh.dims"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# print all years \n",
+ "np.unique(dsh.time.dt.year) #-- or\n",
+ "#pd.Series(dsh.time.dt.year.values).unique()"
]
},
{
@@ -883,7 +835,8 @@
"source": [
"\n",
"### Compute the area weights\n",
- "To account for the different grid cell sizes, we calculate the variable weighted this time.\n"
+ "To account for the different grid cell sizes, we calculate the variable weighted this time.<br>\n",
+ "_**Note**_: It is common practice in climate science to weight grid points by the cosine of their latitude to account for the varying area of grid cells with latitude."
]
},
{
@@ -892,23 +845,29 @@
"metadata": {},
"outputs": [],
"source": [
- "weights = np.cos(np.deg2rad(dsh.lat))"
+ "weights = np.cos(np.deg2rad(dsh.lat))\n",
+ "weights.shape"
]
},
{
- "cell_type": "code",
- "execution_count": null,
+ "cell_type": "markdown",
"metadata": {},
- "outputs": [],
"source": [
- "weights"
+ "### Compute the weighted data."
]
},
{
- "cell_type": "markdown",
- "metadata": {},
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {
+ "tags": []
+ },
+ "outputs": [],
"source": [
- "### Compute the weighted data."
+ "# ANNOTATION: weights.shape (96,)\n",
+ "# clim_xr.tas.shape (12, 96, 192)\n",
+ "# .weighted() applies the weights along the lat dimension\n",
+ "# to all the data in the DataArray -->broadcasting"
]
},
{
@@ -917,18 +876,16 @@
"metadata": {},
"outputs": [],
"source": [
- "clim_xr_wgt = clim_xr.weighted(weights)"
+ "clim_xr"
]
},
{
"cell_type": "code",
"execution_count": null,
- "metadata": {
- "tags": []
- },
+ "metadata": {},
"outputs": [],
"source": [
- "clim_xr_wgt"
+ "clim_xr_wgt = clim_xr.weighted(weights)"
]
},
{
@@ -941,14 +898,14 @@
"source": [
"#-- The weighted call returns a class xarray.core.weighted.DatasetWeighted(obj, weights).\n",
"#-- To obtain the weights as xarray DataArray, use clim_xr_wgt.weights\n",
- "clim_xr_wgt.weights.head(5).values #-- head(5) shows the first 5 values, only."
+ "clim_xr_wgt.weights.head(5).values "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
- "### Compute the spatial mean"
+ "### Compute the area-weighted spatial mean"
]
},
{
@@ -984,7 +941,7 @@
"metadata": {},
"outputs": [],
"source": [
- "clim_xr_mean.tas.plot();"
+ "clim_xr_mean.tas.plot(color='red',marker='o'); "
]
},
{
@@ -1035,7 +992,17 @@
"metadata": {},
"outputs": [],
"source": [
- "anom_xr = tas_xr_wgt.groupby('time.month') - clim_xr_mean"
+ "anom_xr = tas_xr_wgt.groupby('time.month') - clim_xr_mean\n",
+ "# ANNOTATION: clim_xr_mean is area-mean multiyear monthly mean"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "anom_xr"
]
},
{
@@ -1107,7 +1074,7 @@
"pygments_lexer": "ipython3",
"version": "3.10.10"
},
- "toc-autonumbering": true,
+ "toc-autonumbering": false,
"toc-showcode": true
},
"nbformat": 4,
diff --git a/notebooks/xarray_introduction_part1.ipynb b/notebooks/xarray_introduction_part1.ipynb
deleted file mode 100644
index c3783bcc0438b73cc7ce3ced66b9c7a624cd145d..0000000000000000000000000000000000000000
--- a/notebooks/xarray_introduction_part1.ipynb
+++ /dev/null
@@ -1,2664 +0,0 @@
-{
- "cells": [
- {
- "cell_type": "markdown",
- "id": "08a8d53e-167a-480b-beac-15bc6b378f94",
- "metadata": {},
- "source": [
- "***\n",
- "<p align=\"right\">\n",
- " <img src=\"https://www.dkrz.de/@@site-logo/dkrz.svg\" width=\"12%\" align=\"right\" title=\"DKRZlogo\" hspace=\"20\">\n",
- " <img src=\"https://wr.informatik.uni-hamburg.de/_media/logo.png\" width=\"12%\" align=\"right\" title=\"UHHLogo\">\n",
- "</p>\n",
- "<div style=\"font-size: 20px\" align=\"center\"><b> Python Course for Geoscientists, 5-8 March 2024</b></div>\n",
- "<div style=\"font-size: 15px\" align=\"center\">\n",
- " <b>see also <a href=\"https://gitlab.dkrz.de/pythoncourse/material\">https://gitlab.dkrz.de/pythoncourse/material</a></b>\n",
- "</div>\n",
- "\n",
- "***"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "7128b107-2215-4d84-a411-8e0e42fb9a87",
- "metadata": {},
- "source": [
- "\n",
- "<p align=\"center\">\n",
- " <img src=\"https://docs.xarray.dev/en/stable/_static/Xarray_Logo_RGB_Final.svg\" width=\"35%\" align=\"right\" title=\"Xarraylogo\" hspace=\"20\">\n",
- "</p>\n",
- "\n",
- "<font size=\"20\"> Xarray Introduction I</font> \n",
- "\n",
- "Xarray home page: https://xarray.pydata.org/en/stable/index.html <br>\n",
- "Xarray documentation: https://docs.xarray.dev/en/stable/index.html\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "e81e31bd-f599-448f-93b8-8017e91133ae",
- "metadata": {
- "slideshow": {
- "slide_type": "slide"
- },
- "tags": []
- },
- "source": [
- "\n",
- "**Xarray** is a python package which allows us to handle multi-dimensional datasets in a simple way. It provides a huge set of functions for advanced analytics and visualization.\n",
- "\n",
- "**Xarray’s** underlying data model is borrowed from the data format [netCDF](http://www.unidata.ucar.edu/software/netcdf). This data format in combination with the [Climate and Forecast metadata conventions](https://cfconventions.org/) (CF) is the standard for the climate science community. A large part of DKRZ’s data is available in netCDF. Therefore, **Xarray** allows fast and intuitive data analysis on this kind of data, but file formats like GRIB, HDF5, and Zarr can also be used.\n",
- "\n",
- "**Xarray** data structure deals with scientific data by using **dimensions**, **coordinates**, **labels** and **attributes** and extend the capabilities of **NumPy** and **Pandas**.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "6e5188ab",
- "metadata": {
- "slideshow": {
- "slide_type": "slide"
- }
- },
- "source": [
- "# <u>Overview</u>: \n",
- "\n",
- "## Xarray's data model\n",
- "\n",
- "A **data model** describes how the elements of data are organized and standardizes how they relate to one another. \n",
- "**Xarray's** data model consists of the classes **DataArray**, **Dimension**, **Coordinate**, **Dataset** and **Attributes**. \n",
- "(see also https://tutorial.xarray.dev/fundamentals/01_datastructures.html)\n",
- "\n",
- "\n",
- "----\n",
- "**DataArray**: \n",
- "\n",
- "DataArray in xarray is essentially a NumPy array enhanced with additional information such as dimension names, coordinates, and attributes. \n",
- "A DataArray in xarray is like a data variable in a netCDF file\n",
- "\n",
- "----\n",
- "**Dimensions**: \n",
- "\n",
- "Named dimension axes. If no dimension names are defined, then they are named dim_0, dim_1, ... by default.\n",
- "\n",
- "----\n",
- "**Coordinates**: \n",
- "\n",
- "An array which labels a dimension. Two types are defined \n",
- "a) dimension coordinates - 1-dimensional coordinate array assigned to the DataArray with a name and dimension name. \n",
- "b) non-dimensional coordinate - a coordinate array assigned to DataArray with the name assigned to the coordinates and not to the dimensions.\n",
- "\n",
- "----\n",
- "**Dataset** ( dataset or file ): \n",
- "\n",
- "Dictionary-like collection of DataArray objects with aligned dimensions. Similar use of variables, dimensions, coordinates, and attributes like for DataArray. You can see an xarray Dataset as a netCDF file like object. \n",
- "\n",
- "----\n",
- "**Attributes**: \n",
- "\n",
- "Xarray allows you to attach metadata and attributes to both DataArrays and Datasets. \n",
- "Metadata can include information about units, descriptions, and any other relevant information about the data.\n",
- "\n",
- "----\n",
- "\n",
- "\n",
- "<br>\n",
- "\n",
- "<img src=\"https://storage.googleapis.com/jnl-up-j-jors-files/journals/1/articles/148/submission/proof/148-10-1829-1-17-20170405.png\" alt=\"xarray data structure\" border=1 width=900></img> \n",
- "<figcaption align = \"center\"> An overview of xarray’s main data structures. From Hoyer and Hamman (2017); DOI: 10.5334/jors.148 </figcaption>\n",
- "<br>"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "16667921-d50c-459b-a250-7cbc10846a8a",
- "metadata": {},
- "source": [
- "## Dimensionality of arrays\n",
- "\n",
- "Python is **'row major'** which means that the `left dimension varies slowest` and the `right dimension varies fastest`. That's the case why the geo-referenced data have often the dimension order (time, level, lat, lon)."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "f1c5892f-918b-4c2e-8880-df278c6485bb",
- "metadata": {},
- "source": [
- "# xarray DataArrays and Datasets"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "d5bf9dc7",
- "metadata": {
- "slideshow": {
- "slide_type": "slide"
- }
- },
- "source": [
- "## Importing modules\n",
- "\n",
- "In this notebook we work with the Python libraries NumPy, Pandas, Xarray and cfgrib. \n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "79240aa7",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "outputs": [],
- "source": [
- "import xarray as xr\n",
- "import numpy as np\n",
- "import pandas as pd\n",
- "try:\n",
- " import cfgrib\n",
- "except ImportError:\n",
- " import subprocess\n",
- " subprocess.run([\"bash\", \"-c\", \"pip install --user ecmwflibs --quiet\"])\n",
- "\n",
- "from datetime import datetime\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "c4fc03a0-d544-4e6d-aefc-9e66e5b010db",
- "metadata": {},
- "source": [
- "## DataArray\n",
- "\n",
- "The `DataArray` of **Xarray** is the implementation of a labeled multi-dimensional array.\n",
- "\n",
- "To see what this means, we start with the creation of a simple DataArray that is based on an NumPy ndarray.\n",
- "\n",
- "Create NumPy _ndarray_ with shape(4,5):"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "3744f16b-3b33-4fd2-a308-45ec4e121db4",
- "metadata": {},
- "outputs": [],
- "source": [
- "array = np.arange(1,21).reshape(4,5)\n",
- "array"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "383fabdc-978c-45ed-ad1e-c4bd3c98d8ad",
- "metadata": {},
- "source": [
- "Now, we can use the function `xr.DataArray()` to create a DataArray from the NumPy array above."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "986cd1aa-d4d8-44cb-91a5-00ecdae30612",
- "metadata": {},
- "outputs": [],
- "source": [
- "da = xr.DataArray(array)\n",
- "da"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "f9bbb85d-55c4-4807-bf51-0e43fadcb18f",
- "metadata": {},
- "source": [
- "As you can see, the `xr.DataArray()` adds two dimensions named **dim_0** and **dim_1** to the new data array structure. When the function `xr.DataArray()` is used, it returns a data object with some presettings like Coordinates, Indexes and Attributes. In our case these are empty because we did not declared them yet. You can either add them in the `xr.DataArray()` function call or afterwards.\n",
- "Also, you can specify the name of the dimensions when creating the DataArray with `xr.DaraArray` or afterwards using the `rename` method. Note: `rename` returns a new DataArray object."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "e433b8dc-704b-48ab-841f-8d6f03d403ba",
- "metadata": {},
- "outputs": [],
- "source": [
- "da = da.rename({'dim_0':'y','dim_1':'x'})\n",
- "da"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "81fc704b-72e0-472d-832e-e13e5731b6b6",
- "metadata": {},
- "source": [
- "In the next step we assign the arrays x and y which we want to use as coordinates for our DataArray."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "06c9b6d7-4902-454e-aaf6-ea8c0bd3f260",
- "metadata": {},
- "outputs": [],
- "source": [
- "x = np.arange(0., 21., 5.)\n",
- "y = np.arange(0., 20., 5.)\n",
- "\n",
- "print(x, y)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "e3e071e7-e5f4-40ea-a1ac-c79bfc171c51",
- "metadata": {},
- "source": [
- "Xarrays allows us to do the following steps within one `xr.DataArray()` call:\n",
- "\n",
- "- the first dimension should be 'y' and the second 'x'\n",
- "- use the same names as for dims for the coords\n",
- "- assign values to the coords\n",
- "- define the attribute 'standard_name', see https://cfconventions.org/Data/cf-standard-names/current/build/cf-standard-name-table.html; \n",
- " we assume that our DataArray represents a variable with the the standard_name 'age_of_sea_ice' "
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "fbfdd85c-2e60-4bb6-9250-fed8e781a073",
- "metadata": {},
- "outputs": [],
- "source": [
- "da = xr.DataArray(array, \n",
- " dims=('y','x'), \n",
- " coords={'y': y, 'x': x},\n",
- " attrs={'standard_name':'age_of_sea_ice'})\n",
- "da"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "532664b1-1317-40de-8893-f64f8a46443b",
- "metadata": {},
- "source": [
- "It is also possible to name the DataArray itself, e.g. 'var'. You can set it when the DataArray is defined or you can add it later."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "9a8cdbc8-92e1-4026-8b07-fd378c15149d",
- "metadata": {},
- "outputs": [],
- "source": [
- "da = xr.DataArray(array, \n",
- " name='var',\n",
- " dims=('y','x'), \n",
- " coords={'y': y, 'x': x},\n",
- " attrs={'standard_name':'age_of_sea_ice'})\n",
- "da"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "5d2ef647-dcb7-41dd-a397-e6df806f3236",
- "metadata": {},
- "source": [
- "Change the DataArray name of an already existing DataArray to 'var2'."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "467cc1cc-65a2-4875-9611-22f946c55ce8",
- "metadata": {},
- "outputs": [],
- "source": [
- "da.name = 'var2'\n",
- "\n",
- "#print(da)\n",
- "da"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "9a67740a-7073-4253-a933-ddae31b5635b",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "To add another attribute to the DataArray use attrs, for instance set the units attribute."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "4acdb9b1-d10b-4f4e-8a32-45bafd5c1f98",
- "metadata": {},
- "outputs": [],
- "source": [
- "da.attrs['units'] = 'year'\n",
- "\n",
- "da"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "346d20bc-c44d-4732-a2dd-b6b21507b71e",
- "metadata": {},
- "source": [
- "### Expand dimensions\n",
- "\n",
- "You can add a dimension, e.g. time, to the already existing DataArray with `DataArray.expand_dims()`. In the next example, we add a time dimension of length 2 with values 1 and 2 to our DataArray."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "150fdf6d-770d-4741-8a29-ca4ca3db2b69",
- "metadata": {},
- "outputs": [],
- "source": [
- "time = [1,2]\n",
- "\n",
- "da.expand_dims({'time':time}, axis=0)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "8c8aa296-5fcb-49fa-8309-bbe65f3e303d",
- "metadata": {},
- "source": [
- "The time dimension and its data is added to the DataArray but as we can see the data array itself is duplicated. This is caused by the fact that our input data **array** is of shape(4,5) (which can be reshaped into (1,4,5)) but now it has the shape(2,4,5). The _missing_ data for the second time step is copied from the first timestep."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "6b38c69c-e6fe-4e06-a8f0-f27f67b333d3",
- "metadata": {},
- "source": [
- "Note that the DataArray.expand_dims() just **returns** a DataArray with this new dimension, it does not replace it."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "417da472-bdd0-4857-a29a-3f122ad23ee9",
- "metadata": {},
- "outputs": [],
- "source": [
- "da"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "3bf92ff6-df4b-41c2-a646-bbd2cd74d24c",
- "metadata": {},
- "source": [
- "We therefore update our variable _da_ by assigning the returned DataArray."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "f2202f0b-dec4-4078-abc3-a8d080beda0b",
- "metadata": {},
- "outputs": [],
- "source": [
- "da = da.expand_dims({'time':time}, axis=0)\n",
- "\n",
- "da"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "3f1250bc-3a03-4c92-b207-435a903289df",
- "metadata": {},
- "source": [
- "To retrieve the shape of the DataArray use the shape or size property."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "0f702241-0ca8-469a-9fc8-355ec8b53298",
- "metadata": {},
- "outputs": [],
- "source": [
- "da.shape"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "2b1ea8c1-516b-4885-9445-159b554695ac",
- "metadata": {},
- "outputs": [],
- "source": [
- "da.sizes"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "3a300235-4584-4722-b654-97a76b571082",
- "metadata": {},
- "source": [
- "The result shows that we now have two time steps and a 3-dimensional array with the dimensions time, y and x while our input data, before expand_dims, was a 2-dimensional array. "
- ]
- },
- {
- "cell_type": "markdown",
- "id": "05975516-b469-43dd-b84f-5df1bbe3bd05",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "----\n",
- "\n",
- "# Exercises:"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "a3fe1395-c020-4203-a77d-27839a2ec49d",
- "metadata": {},
- "source": [
- "## Exercise: `xr.DataArray`\n",
- "\n",
- " Make yourself familiar with `xr.DataArray`\n",
- "1. generate an Xarray DataArray\n",
- "2. add some attributes, including a standard_name attribute\n",
- "3. change the default dimension names and add coordinate values\n",
- "4. create the same DataArray with just one call of xr.DataArray"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "ed37edda-48b4-4447-954f-e0779b19dd11",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "bcd00fc9-eec6-4341-baf4-5b1fa5ea66d6",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 2. \n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "04d2be73-0c53-4cd9-929d-ebd0a3a153d9",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 3. \n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "b3492698-e9e4-460a-bc2a-29c13dbf320d",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 4.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "b3bd0f5b-8e7e-4aed-963d-fc36154430ac",
- "metadata": {
- "tags": []
- },
- "source": [
- "<br>\n",
- "\n",
- "### Solution\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "06078ba7-53e5-489c-8f0c-8c6b41168e6c",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1. generate an Xarray DataArray\n",
- "\n",
- "np.random.seed(100000)\n",
- "nt = 5\n",
- "\n",
- "data = xr.DataArray(np.random.random((nt,4,5)))"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "10c8e399-6de1-40c9-a8b0-b72237e477ee",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 2. add some attributes\n",
- "\n",
- "data.attrs['my_attr'] = 'my new attribute'\n",
- "data.attrs['creation_date'] = datetime.today().strftime('%Y-%m-%d')"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "d8f2b110-b5fe-42fb-87a3-7768c20f5c65",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 3. change the default dimension names and add coordinate values\n",
- "\n",
- "data = data.rename({'dim_0':'t', 'dim_1':'y', 'dim_2':'x'})"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "59c1b03d-b2bc-482d-a86e-58e4597ded2d",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 4. create the same DataArray with just one call of xr.DataArray\n",
- "\n",
- "nt = 5\n",
- "np.random.seed(100000)\n",
- "data = np.random.random((nt,4,5)) * 2000\n",
- "\n",
- "data_xr = xr.DataArray(data, \n",
- " dims=('index', 'axis_x','axis_y'), \n",
- " coords={'index': np.arange(1,nt+1), \n",
- " 'axis_x': [2, 4, 6, 8], \n",
- " 'axis_y': [1,2,3,4,5]},\n",
- " attrs={'standard_name':'fire_temperature',\n",
- " 'units':'K', \n",
- " 'comment': 'Random data min=0., max=2000.'})\n",
- "#data_xr"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "2743d4dd",
- "metadata": {
- "slideshow": {
- "slide_type": "slide"
- },
- "tags": []
- },
- "source": [
- "<br>\n",
- "\n",
- "----\n",
- "\n",
- "## More about DataArrays\n",
- "\n",
- "Let's first compare a NumPy array with a Xarray DataArray. You can directly convert a NumPy array into an Xarray DataArray type by using it as input for Xarray's function `xr.DataArray`. We use the _atmosphere water vapor content_ data from the file `../data/prw.dat` by loading it with NumPy.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "132c2c56-91cc-4ac6-a245-cabe5838f48f",
- "metadata": {},
- "source": [
- "Show the first 5 lines of the ascii input file"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "56fb99e3",
- "metadata": {},
- "outputs": [],
- "source": [
- "!head -5 ../data/prw.dat"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "8442edcb-3150-4f45-aba5-ddc30d094097",
- "metadata": {},
- "source": [
- "Read columns 1 to 3 of the input file while skipping the header"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "cc5e297c-18be-480b-acd5-251c02487248",
- "metadata": {},
- "outputs": [],
- "source": [
- "prw_data = np.loadtxt('../data/prw.dat', usecols=(1,2,3), skiprows=1)\n",
- "prw_data"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "176c65ae-ecc4-4888-b842-5242c38dd753",
- "metadata": {},
- "source": [
- "Convert the numpy array into an Xarray DataArray"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "9072d2cf-9f24-4d68-bcfa-cc923e22d06b",
- "metadata": {},
- "outputs": [],
- "source": [
- "prw_data_xr = xr.DataArray(prw_data)\n",
- "prw_data_xr"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "cf189b78-b491-4149-9b37-15987d189489",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "prw_data_xr.attrs"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "df931aaf",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "`prw_data_xr` has got more structure and descriptive information than `prw_data`. In contrast to the `NumPy` data array, the `Xarray` DataArray can separate the variable of interest, `prw`, as a *data variable* from *coordinate* variables since the `Xarray` DataArray has the Classes:\n",
- "\n",
- "- **dimensions** with names (`prw_data_xr.dims`)\n",
- "- **coordinates** pointing to variables (`prw_data_xr.coords`)\n",
- "- **attributes** (`prw_data_xr.attrs`)\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "7cdba197",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "This information is not correctly parsed from the input NumPy array when executing `xr.DataArray()`, but we configure them in the call `xr.DataArray()` via the function parameters (arguments + keyword arguments):\n",
- "\n",
- "```python\n",
- "xr.DataArray(data,\n",
- " coords=,\n",
- " dims=,\n",
- " name=,\n",
- " attrs=,\n",
- " )\n",
- "```"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "7444d69a",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "<div class=\"alert alert-info\">\n",
- " <b>Note:</b> When working with <b>xarray</b>, the arguments and keyword arguments for a function are <i>in general</i> very usefull and important!\n",
- "</div>"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "d155a99a-ac6e-4e6b-91fd-a6abc37c6082",
- "metadata": {},
- "source": [
- "The configuration of coordinate values is not only important for `Xarray` but also other software tools since **labeled geospatial** information from coordinates is required, e.g. for\n",
- "\n",
- "- **plotting**: mapping of data on a real world grid point\n",
- "- **analysis**: routines e.g. calculating area **weighted means**\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "a73b863b",
- "metadata": {
- "slideshow": {
- "slide_type": "slide"
- }
- },
- "source": [
- "### Parsing NumPy data with labels to xarray\n",
- "\n",
- "Let's define a clear structure for the `xarray.DataArray()` for the NumPy data first:\n",
- "\n",
- "1. The actual **data** for the data variable is in the first column of the NumPy array.\n",
- "2. The **coords** are the second and third column of the NumPy array. They have the same dimension as the data array.\n",
- "3. We have one dimension (**dims**) which refers to the **_station_**. It is an index which runs from 0 to the length of the a column minus 1.\n",
- "4. The **name** of the data variable is **prw**.\n",
- "5. In the **attrs**, we can store variable attributes like **_units_**. The **standard_name** of prw is **_atmosphere_mass_content_of_water_vapor_**; the corresponding canonical units is **_kg m-2_**.\n",
- "\n",
- "Let's bring that into context with `xr.DataArray()`:"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "bdd57e5c",
- "metadata": {},
- "outputs": [],
- "source": [
- "prw_data_xr = xr.DataArray(prw_data[:,2],\n",
- " coords={\"lat\":(\"Station\", prw_data[:,0]),\n",
- " \"lon\":(\"Station\", prw_data[:,1])},\n",
- " dims=[\"Station\"],\n",
- " name=\"prw\",\n",
- " attrs={\"units\":\"kg m-2\",\n",
- " \"standard_name\":\"atmosphere_mass_content_of_water_vapor\"})\n",
- "prw_data_xr"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "de946f3f",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "outputs": [],
- "source": [
- "print(\"Variable Name: \", prw_data_xr.name)\n",
- "print(\"Dimensions: \", prw_data_xr.dims)\n",
- "print(\"Coordinates: \", prw_data_xr.coords)\n",
- "print(\"Sizes: \", prw_data_xr.sizes)\n",
- "print(\"Attribute: \", prw_data_xr.attrs)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "c32d2480",
- "metadata": {
- "slideshow": {
- "slide_type": "slide"
- }
- },
- "source": [
- "### Dimensions\n",
- "\n",
- "Dimensions are **indices** covering an interval of the length of the dimension.\n",
- "\n",
- "In our example, we only have one dimension where each index refers to one **station**. However, if we create a quick plot of the data with the function `xr.DataArray.plot()`, we only get a one dimensional view:"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "206c9984",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "outputs": [],
- "source": [
- "prw_data_xr.plot();"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "1f9d1a71-cfec-4402-942e-27dde0d58102",
- "metadata": {
- "slideshow": {
- "slide_type": "slide"
- }
- },
- "source": [
- "\n",
- "## Exercise\n",
- "\n",
- "1. Create a two dimensional NumPy with the size `len(prw_data)` x `len(prw_data)`\n",
- "\n",
- "1. Assign `NaN` values to the entire array\n",
- "\n",
- "1. On the diagonal of the quadratic array, insert the values of `prw_data`\n",
- "\n",
- "1. Show the new data frame\n",
- "\n",
- "<br>\n",
- "\n",
- "You will need:\n",
- "\n",
- "- `np.full()` or `np.empty()`\n",
- "- `np.Nan`\n",
- "- use a `for` loop\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "e0717073",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "outputs": [],
- "source": [
- "# 1.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "ce710f31-2f89-4a0a-9a67-24a725f01103",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 2.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "4baf6447-377d-4abb-88ac-4dbc55efa73c",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 3.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "cb219687-61e8-48e3-9dc6-41af54e0618e",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 4.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "f75d2261-bf09-4f48-b3fa-4ebe004a5b05",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "### Solution"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "a071554f-ff6a-4fd5-bd81-8ecf8b33a895",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1. and 2.\n",
- "prw_data_2d = np.full([len(prw_data),len(prw_data)], np.nan)\n",
- "\n",
- "# another way to generate the prw_data_2d\n",
- "#prw_data_2d = np.empty([len(prw_data),len(prw_data)]) * np.nan"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "daf21374-4bb0-4305-9479-853d90b422f3",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 3. \n",
- "for i in range(0, len(prw_data)):\n",
- " prw_data_2d[i,i] = prw_data[i,2]\n",
- " \n",
- "print(prw_data_2d)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "b76a650e-8fd4-4249-9e5a-d166eefe294c",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 4. \n",
- "xr.DataArray(prw_data_2d).plot()"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "211d4eb6",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "<br>\n",
- "\n",
- "# Exercise\n",
- "Let's pass this DataArray to **Xarray**.\n",
- "\n",
- "1. Reset the variable `pwr_data_xr` with a `xr.DataArray()` but use `prw_data_2d` as input.<br>\n",
- " **Hint**: Set the dims to \\[\"lat\",\"lon\"\\]. Coordinate and dimension names have to be the same.\n",
- "2. Plot again\n",
- "\n",
- "<br>\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "c6a5954b",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "outputs": [],
- "source": [
- "# 1.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "f7a69330-b974-476d-b12f-4c011da36f9d",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 2.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "cad81b75-81aa-44bc-90d9-44392046df2a",
- "metadata": {
- "tags": []
- },
- "source": [
- "<br>\n",
- "\n",
- "\n",
- "### Solution"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "0e51ca28-cf0d-4e06-b5f8-32e834246aeb",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 1.\n",
- "\n",
- "prw_data_xr = xr.DataArray(prw_data_2d,\n",
- " coords={\"lat\": prw_data[:,0],\n",
- " \"lon\": prw_data[:,1]},\n",
- " dims=[\"lat\",\"lon\"],\n",
- " name=\"prw\",\n",
- " attrs={\"units\":\"kg m-2\",\n",
- " \"standard_name\":\"atmosphere_mass_content_of_water_vapor\"})\n",
- "print(prw_data_xr)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "367b4688-5099-4566-90f4-d7c0e21ce5e9",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 2. his leads to an error (please uncomment the following line for testing)\n",
- "\n",
- "#prw_data_xr.plot()"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "79102445",
- "metadata": {
- "slideshow": {
- "slide_type": "slide"
- },
- "tags": []
- },
- "source": [
- "An error occurs due to the fact that we have station data that do not have ascending or descending coordinate values. The **ValueError** at the end of the error message gives you the hint to use `sortby` to solve the error.\n",
- "\n",
- "The plot only uses the indices of the dimensions for the x and y axes of the plot. This is because the **coordinates** 'lat' and 'lon' are not interpreted as **_index coordinates_**. **Xarray** will interpete coordinates as **index coordinates** only if the name of the coordinate is the same as the name of the dimension. "
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "474826b0-c5bc-4e12-a6ae-0e4774127cd0",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 2. correct way\n",
- "\n",
- "prw_data_xr = prw_data_xr.sortby(['lon','lat'])\n",
- "prw_data_xr.plot()\n",
- "\n",
- "\n",
- "print(prw_data_xr.lon.min().data, prw_data_xr.lon.max().data)\n",
- "print(prw_data_xr.lat.min().data, prw_data_xr.lat.max().data)\n",
- "print(prw_data_xr.min().data, prw_data_xr.max().data)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "636f7b70",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "<br>\n",
- "\n",
- "We created a simple plot which gives us an idea of for which locations we have valid station data using only few **Xarray** commands. In the session Visualization Part 2., we will learn a more sophisticated plotting including e.g. *coastlines*.\n",
- "\n",
- "Here is a taste:\n",
- "\n",
- "<br>"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "620c0585",
- "metadata": {
- "slideshow": {
- "slide_type": "skip"
- }
- },
- "outputs": [],
- "source": [
- "import cartopy.crs as ccrs\n",
- "import matplotlib.pyplot as plt\n",
- "\n",
- "proj = ccrs.PlateCarree() # choose map projection\n",
- "\n",
- "fig, ax = plt.subplots(figsize=(12,8), subplot_kw={'projection':proj})\n",
- "ax.set_extent([-105, -80, 25, 41], proj)\n",
- "ax.stock_img() # add satellite image\n",
- "ax.gridlines(draw_labels=True, color='None', zorder=0) # turn on axis label, turn off gridlines\n",
- "ax.coastlines() # add coastlines\n",
- "\n",
- "prw_data_xr.plot(cmap='Reds', cbar_kwargs=dict(shrink=0.6)) ; # decrease colorbar size"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "1956f70a",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "## Exercise\n",
- "\n",
- "Play a bit with the Xarray DataArray and use the DataArray **prw_data_xr** from above\n",
- "\n",
- "1. add a variable long_name attribute \n",
- " (name it as you like, but be aware that many plotting routines parse the long_name to plot the labels ;))\n",
- "2. change the standard_name and variable name\n",
- "3. add more attributes and print them all\n",
- "\n",
- "<br>"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "33b94877",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "f6e4fee9-1189-45ea-bebb-a64479810e20",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 2.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "df5a1a73-a371-4a2e-9755-a5a552e46c98",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 3.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "bf0c4d90-8058-4828-b5d6-195ac6b89ce7",
- "metadata": {
- "tags": []
- },
- "source": [
- "<br>\n",
- "\n",
- "### Solution"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "56a48e8e-22dc-41f0-b9a8-95544dc10a08",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 1.\n",
- "prw_data_xr.attrs['long_name'] = 'This is the variables long_name'"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "3b3072d1-0209-4246-8ac7-5f463b399def",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 2.\n",
- "prw_data_xr['standard_name'] = 'area_fraction'\n",
- "prw_data_xr.name = 'variable_A'"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "cd30d47e-98da-40ad-9fb5-4f08b49e7515",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 3. \n",
- "prw_data_xr.attrs['created_by'] = 'DKRZ Python Course'\n",
- "prw_data_xr.attrs"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "c7b077fa-2b15-49d4-8ee2-f9845b7d1836",
- "metadata": {},
- "source": [
- "----\n",
- "## Dataset\n",
- "\n",
- "An Xarray `Dataset` is a dictionairy-like container of data arrays with aligned dimensions. <br><br>\n",
- "\n",
- "\n",
- "\n",
- "Datasets have four key properties:\n",
- "\n",
- " 1. dims: dict for dimension names\n",
- " 2. data_vars: dict of data arrays\n",
- " 3. coords: dict of coordinates\n",
- " 4. attrs: dict for dataset (global) attributes\n",
- "\n",
- "**Note:** <br>\n",
- "If you are familiar with the **netCDF file format**: the Xarray Dataset is designed as an in-memory representation of the netCDF data model."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "c7584b1f-2603-4a1d-8ea2-69d9576c006e",
- "metadata": {},
- "source": [
- "You can use the already defined DataArrays to create a Dataset. Here, we use our prw_data_xr DataArray."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "729203d4-5d77-45b9-bc20-25a81277567c",
- "metadata": {},
- "outputs": [],
- "source": [
- "prw_data_xr"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "d665a100-55b9-4abe-885a-a5a0f0a1bc5f",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds = prw_data_xr.to_dataset()\n",
- "ds"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "7733a4c1-430e-4925-9858-ebc198f0ebf6",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds = prw_data_xr.to_dataset(promote_attrs=True)\n",
- "ds"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "e237fb2e-4722-4f6e-a683-47255f1a22e1",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "Next, we use a NumPy DataArray of random values as input data for the Dataset.\n",
- "\n",
- "1. define two arrays for the variables temp and prec\n",
- "1. define the coordinate data for lat and lon\n",
- "1. define the coordinate data for a time dimension\n",
- "1. create the Dataset\n",
- "\n",
- "<br>"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "9245b20d-b999-48b1-b1d5-bdb577778865",
- "metadata": {},
- "source": [
- "1a. Define the data for the variable temp (temperature)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "5c5a2ca5-3e7b-4885-a5d5-01136829fe26",
- "metadata": {},
- "outputs": [],
- "source": [
- "temp = np.random.uniform(250,300,40).reshape((2,4,5))\n",
- "temp"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "57928a51-54fc-48ba-868f-85848eebc2ba",
- "metadata": {},
- "source": [
- "1b. Define random data for a variable prec (precipitation), for reproducability we set the random seed."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "c57eaccd-8553-4f3c-aa10-6cdeeb768c70",
- "metadata": {},
- "outputs": [],
- "source": [
- "np.random.seed(100000)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "0b7d2b20-2ce5-4c2a-98b9-5db8f34ed651",
- "metadata": {},
- "outputs": [],
- "source": [
- "prec = np.random.uniform(0.001,0.015,40).reshape((2,4,5))\n",
- "\n",
- "prec"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "4d6b2a53-c1e3-4005-b5b9-fcb44775699d",
- "metadata": {},
- "source": [
- "2. Define the data for the coordinate variables lat and lon"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "d3e75711-3144-41ed-8e26-89610bb23125",
- "metadata": {},
- "outputs": [],
- "source": [
- "lat = [45.,50.,55.,60.]\n",
- "lon = [0.,5.,10.,15.,20.]"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "de9ed707-0113-4b33-83c1-3ae403d1eccd",
- "metadata": {},
- "source": [
- "3. Define the time variable\n",
- "\n",
- "This time we generate a time variable containing 2 time steps with daily-frequency with **Pandas** `pd.date_range()` function."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "c9741f48-098b-4910-b62c-6542f3f7f479",
- "metadata": {},
- "outputs": [],
- "source": [
- "time = pd.date_range(start='2023-01-01', periods=2)\n",
- "time"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "101878c3-5053-46a2-9698-7be0d18d6fe1",
- "metadata": {},
- "source": [
- "4. Define the Dataset\n",
- "\n",
- "Use the data and coordinate variables to generate the Dataset. Add an attribute 'comment' to the Dataset."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "69472a09-4fc8-47d2-adb2-3d1d7aa857b7",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds = xr.Dataset({'temp': (['time','lat','lon'], temp),\n",
- " 'prec': (['time','lat','lon'], prec),\n",
- " },\n",
- " coords={'time': time,\n",
- " 'lat': (['lat'], lat),\n",
- " 'lon': (['lon'], lon),\n",
- " },\n",
- " attrs={'comment': 'This is a global attribute of the dataset'})"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "bdf6c064-fc29-4077-8ea1-6a92701fe082",
- "metadata": {},
- "source": [
- "Let's look at the created Xarray Dataset"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "2b7a98d4-62c6-4083-9fb8-eae9baeb6f52",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "79c31337-bc9d-4803-b1bf-a6502c237674",
- "metadata": {},
- "source": [
- "If you want to have it more 'ncdump'-like view, use `Dataset.info()`."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "22e3f347-6ab1-433a-8914-25b0f009149f",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds.info()"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "57830e7d-c216-4f82-be96-a8024de57782",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "**Let's access the data.**"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "fd989ce1-908c-4827-a2c9-4e065ce0fc3d",
- "metadata": {},
- "source": [
- "_the data variable temp_"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "a27a538c-ff92-4dfa-9473-91d00a9daed6",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds['temp'] \n",
- "# alternatively:\n",
- "#ds.temp"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "fc452fe4-6416-48c7-8578-1f6e80072752",
- "metadata": {},
- "source": [
- "_the coordinate variable lat_"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "c4603077-e9b4-4a18-8783-5c73cad7247b",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds.lat\n",
- "\n",
- "# you can use the variable coordinate lat, too\n",
- "ds.temp.lat"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "ba3dcc83-7777-4d51-a7cb-664cf48955b2",
- "metadata": {},
- "source": [
- "_the coordinate variable time_"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "385db263-730b-4597-a581-eabb748da8f8",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds.time"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "4a9021fb-d43e-442f-9e7a-d522df387f21",
- "metadata": {},
- "source": [
- "### Dimensions, shape and size\n",
- "\n",
- "To get more information about the dimension, shape and size of a **Dataset**, we can use the appropriate attributes.\n",
- "\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "048e197e-4e8d-4928-87e0-0a4ac93f3b68",
- "metadata": {},
- "outputs": [],
- "source": [
- "dims = ds.dims\n",
- "shape = temp.shape\n",
- "size = temp.size\n",
- "rank = len(shape)\n",
- "\n",
- "print('dimensions: ', dims)\n",
- "print('shape: ', shape)\n",
- "print('size: ', size)\n",
- "print('rank: ', rank)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "45f0a929-e687-4002-9331-cadf545abc2b",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "## Exercise\n",
- "\n",
- "Make yourself familiar with `xr.Dataset`\n",
- "\n",
- "1. generate an Xarray Dataset\n",
- "1. try to add some attributes\n",
- "1. choose a variable and print its content"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "41938413-c665-4e60-af00-b5a6f604f0b8",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "d0ecdf10-d231-4eb6-83c7-f3c37bef6716",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 2.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "89e6a5c3-e54f-46c6-bd4d-9493d25d8038",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 3.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "2a0d1d00-972e-41fd-85f4-f4960d5f4c02",
- "metadata": {
- "tags": []
- },
- "source": [
- "<br>\n",
- "\n",
- "### Solution"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "75c01bb6-8a2d-4dc7-9573-d1109bb14cd9",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 1.\n",
- "tas = xr.DataArray(temp,\n",
- " coords={'time': time,\n",
- " 'lat': (['lat'], lat),\n",
- " 'lon': (['lon'], lon),\n",
- " },\n",
- " name='tas',\n",
- " attrs={'units': 'K', 'standard_name':'surface_temperature'})\n",
- "\n",
- "prc = xr.DataArray(prec,\n",
- " coords={'time': time,\n",
- " 'lat': (['lat'], lat),\n",
- " 'lon': (['lon'], lon),\n",
- " },\n",
- " name='prec',\n",
- " attrs={'units': 'mm', 'standard_name':'precipitation'})\n",
- "\n",
- "\n",
- "ds_new = xr.merge([tas,prc])\n",
- "print(ds_new.tas.attrs)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "9ecc3771-ffcd-44bd-b0b1-1d9a107fda40",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# ... 1. Use NumPy array temp and Xarray DataArray prc\n",
- "\n",
- "ds_new = xr.Dataset({'tas': (['time','lat','lon'], temp.data, {'units':'K', \n",
- " 'standard_name':'surface_temperature'}),\n",
- " 'prc': (['time','lat','lon'], prc.data, prc.attrs),\n",
- " },\n",
- " coords={'time': time,\n",
- " 'lat': (['lat'], lat),\n",
- " 'lon': (['lon'], lon),\n",
- " },\n",
- " attrs={'comment': 'This is a global attribute of the dataset',\n",
- " 'source':'DKRZ Python Course'})\n",
- "print(ds_new.tas.attrs)\n",
- "print(ds_new.prc.attrs)\n",
- "print(ds_new)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "b714cede-5eca-4b39-809e-380d4db44983",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# ... 1. Add a DataArray to an existing Dataset\n",
- "\n",
- "ds_new2 = xr.merge([ds_new, tas.rename('tas2')])\n",
- "ds_new2"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "a45ebe8c-d83b-4fe1-a8fc-f7805b9f094d",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 2.\n",
- "\n",
- "ds_new.tas.attrs['long_name'] = 'near surface temperature'\n",
- "\n",
- "print(ds_new.tas.attrs)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "d0f698a0-69de-4bce-bcaf-0d1a3132dbda",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 3.\n",
- "\n",
- "print(ds_new.prc.data)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "262c991d-617a-4cdf-b981-b851771d0951",
- "metadata": {},
- "source": [
- "# Indexing and slicing data \n",
- "\n",
- "See also: https://docs.xarray.dev/en/stable/user-guide/indexing.html#\n",
- "\n",
- "To demonstrate how to do DataArray indexing we create a small DataArray of shape(3,5). \n",
- "\n",
- "In <u>this example DataArray</u> the dimension **x** can be seen as **row** and the dimension **y** as **columns**."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "dcbd2a1e-9d4f-4678-95fe-2189d3e736cb",
- "metadata": {},
- "outputs": [],
- "source": [
- "da = xr.DataArray(np.arange(1,16).reshape((3,5)),\n",
- " dims=['x', 'y'],\n",
- " coords={'x':[1,2,3], 'y':[10,20,30,40,50]})\n",
- "da"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "66ab6947-6e55-456a-8891-ca4edbb099cd",
- "metadata": {},
- "source": [
- "You can extract data using the indices of the dimensions. There are different ways to extract data from the DataArray.\n",
- "\n",
- "For the DataArray da with 2 dimensions using only one index for the 2d-array means you select a complete 'row' ('x')."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "22f7b17d-8451-41b8-8ba9-bf28bd356282",
- "metadata": {},
- "outputs": [],
- "source": [
- "da[0]"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "3333c285-67c2-42ca-87f1-3bbf919ce34b",
- "metadata": {},
- "source": [
- "When using 2 indices you can extract single values. You can use\n",
- "\n",
- " data_array [index_of_dim_0][index_of_dim_1]\n",
- " \n",
- "or \n",
- " \n",
- " data_array [index_of_dim_0, index_of_dim_1]"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "ea9c66e6-6a05-48d9-835c-0b48ef732661",
- "metadata": {},
- "outputs": [],
- "source": [
- "da[1][0]"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "19bfd49d-8469-49d1-b58f-2f148ec3a129",
- "metadata": {},
- "outputs": [],
- "source": [
- "da[1,0]"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "54ef3f18-18f1-4406-9dc0-5e68fa259359",
- "metadata": {},
- "source": [
- "There is a method for DataArrays and Datasets called .isel() which uses the dimension name and the integer index.\n",
- "\n",
- "The following command does the same as the last example from above."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "98d4c335-da25-4641-a265-2c44b8e4b0a7",
- "metadata": {},
- "outputs": [],
- "source": [
- "da.isel(x=1, y=0)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "099763ea-9a97-4691-8fee-22359616cd6f",
- "metadata": {},
- "source": [
- "So far we have selected only one element but we want now select more and therefor we use the slicing method (as shown in NumPy).\n",
- "\n",
- "Select some 'rows':"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "f47628dc-b33d-4ec2-b18a-e1f049cba88b",
- "metadata": {},
- "outputs": [],
- "source": [
- "da[0:2, 1:3]"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "b21cf277-3e8e-47b5-8bed-9f5dc544d1dc",
- "metadata": {},
- "source": [
- "With the `slice` function you can extract slices from the DataArray using the `.sel()` method."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "4d543732-7b74-44cb-8a51-e26eee99d4a8",
- "metadata": {},
- "outputs": [],
- "source": [
- "da.isel(x=slice(0,2), y=0)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "cd013b19-fc63-4125-bec0-9a067a4355e6",
- "metadata": {},
- "source": [
- "## Label-based indexing\n",
- "\n",
- "Insted of using the index integer value you can also lookup the dimensions by name."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "da06f5ab-5870-4e57-b360-5289c74e740a",
- "metadata": {},
- "outputs": [],
- "source": [
- "da.sel(x=3)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "202de62c-9c10-4d4e-b023-1305970e1534",
- "metadata": {},
- "source": [
- "Do you know what we mean? Let us use a better example next.\n",
- "\n",
- "Therefore, we use the Dataset with the temperature and precipitation variable from above to demonstrate the `.sel()` and `.loc()` methods. Both can also be used with DataArrays."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "d4a0dd89-9de5-4840-a4b9-d348c0a3d8eb",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "c7a1fcd1-fc79-43cd-a3f6-c5bffe897249",
- "metadata": {},
- "source": [
- "Using the .sel() method with a Dataset it has an impact to all data variables (temp, precip).\n",
- "\n",
- "In the next example we want to extract only the data of all variables for time step '2020-01-15'."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "5c9ad0ac-3342-45bb-a88e-4f7c0088d742",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds.sel(time='2023-01-01')"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "63a96f7d-5bf8-40c2-bb02-0eef06f6653c",
- "metadata": {},
- "source": [
- "Extract the temp data of a single time step."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "82b1e95a-d232-4e9b-84d9-804ef9cfd7a1",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds.temp.sel(time='2023-01-01')"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "f3e92f18-e69a-4855-80ce-1766a5ed41b9",
- "metadata": {},
- "source": [
- "You can combine multiple labels at the same time to extract data. If you do not know the exact values you can use the keyword method with nearest to find the dimension index nearest to the given value."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "81115f29-aacd-4f0d-bfc5-f419bac15d90",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds.temp.sel(lat=51.5, lon=2.5, method='nearest').values"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "b48a9c42-7170-47d5-b2df-748a7c603906",
- "metadata": {},
- "source": [
- "Note: The keyword method can't be used with dimension slicing."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "ce489175-c176-41b2-922e-f9080d88c9c6",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds.temp.sel(time='2023-01-01', lat=slice(51.5,57.5)).values"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "e9da4f10-c1f7-469c-8534-4f79d18358d2",
- "metadata": {},
- "source": [
- "If you would prefer to work more Panda-like, then you can use the .loc[] method that uses a dictionary."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "68397391-7fc1-4563-a556-310f2f7ce59c",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds.temp.loc[{'time':'2023-01-01'}]"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "b4ae0c1a-7ef8-46bc-8adf-3db72b5e766e",
- "metadata": {},
- "source": [
- "Overview of the four different kinds of indexing:\n",
- "\n",
- "| Dimension lookup | Index lookup | DataArray syntax | Dataset syntax |\n",
- "|:-----------------|:--------------|:-------------------------------------------------|:-------------------------------------------------|\n",
- "| Positional | By integer | `da[0, :, :]` | not available |\n",
- "| Positional | By label | `da.loc[\"2001-01-01\", :, :]` | not available |\n",
- "| By name | By integer | `da.isel(time=0)` or <br> `da[dict(time=0)]` | `ds.isel(time=0)` or <br> `ds[dict(time=0)`] |\n",
- "| By name | By label | `da.sel(time=\"2001-01-01\")` or <br> `da.loc[dict(time=\"2001-01-01\")`] | `ds.sel(time=\"2001-01-01\"`) or <br> `ds.loc[dict(time=\"2001-01-01\")]` |"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "62498905-f033-4c0c-a28c-3ec1bd6f3b95",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "## Exercise:"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "9c22ebd0-af37-4643-93a7-c7784d79d333",
- "metadata": {},
- "source": [
- "1. Extract some precipitation data from the Dataset ds using\n",
- " - .isel()\n",
- " - .sel()\n",
- " - .loc[]\n",
- "2. Which method do you like better `.sel()` or `.loc[]`?"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "3370f2ba-13c4-4336-afd8-07d06099b77c",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1 - a\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "56992a3b-c3ee-46c3-bd2d-ea6acae788ab",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1 - b\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "9b6f873b-6cb1-40c0-9322-7212d938310c",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1 - c\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "d448b568-6a3a-4896-bda3-cf75c2e3b480",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "---- \n",
- "# Write DataArray or Dataset to file\n",
- "\n",
- "Xarray provides an easy way to write the well defined Dataset to an netCDF file with the function `.to_netcdf()`."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "fa43249d-bf4e-4196-8081-2d78f64a4d6c",
- "metadata": {},
- "outputs": [],
- "source": [
- "!rm ds_output_file.nc\n",
- "\n",
- "ds.to_netcdf('ds_output_file.nc')"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "719e1bdc-33d9-4e7d-b18f-997efb317472",
- "metadata": {},
- "outputs": [],
- "source": [
- "!ncdump -h ds_output_file.nc"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "191abdc2-790e-4c69-acd6-34bc9d22385d",
- "metadata": {},
- "source": [
- "That was really easy! But for completeness we should have added some more attributes to the dimensions and data variables like units, standard_name, and others.\n",
- "\n",
- "Let's see how it looks like when we write the DataArray to a netCDF file."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "47dcefe8-b5b4-467f-b36e-a9e115b1f7ff",
- "metadata": {},
- "outputs": [],
- "source": [
- "!rm da_output_file.nc\n",
- "da.to_netcdf('da_output_file.nc')"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "4f47afa7-c5fe-4add-9995-2828edaf9703",
- "metadata": {},
- "outputs": [],
- "source": [
- "!ncdump -h da_output_file.nc"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "d932f89b-b1fd-4c66-9a47-c4965a768373",
- "metadata": {},
- "source": [
- "It is also possible to write the Dataset to a Zarr file with the `Dataset.to_zarr()` function.\n",
- "\n",
- "Note:\n",
- "To write the data to a CSV file you can convert the Dataset to a `Pandas.DataFrame` and then use the `pandas.DataFrame.to_csv()` function. An alternative is to use the **xarray_extras** package.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "2dcef030-113c-450f-9e0c-2d2e3a5b7086",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "----\n",
- "\n",
- "# Open and read file with xarray\n",
- "\n",
- "In the next step we want to read our newly created netCDF file. Xarray provides the function `xr.open_dataset()` to open a file with the file format netCDF, GRIB, HDF5, or Zarr. Default format is netCDF.\n",
- "\n",
- " ds_in = xr.open_dataset('infile.nc')\n",
- "\n",
- "is the same as\n",
- "\n",
- " ds_in = xr.open_dataset('infile.nc', engine='netCDF4')"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "2bc976b1-809d-4615-af4f-c3e600325119",
- "metadata": {},
- "source": [
- "## Open and read a netCDF file"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "8bf18448-6a9a-468d-8d0e-4133e72a9de8",
- "metadata": {},
- "source": [
- "\n",
- "As the function name says it only opens the file and reads in the meta-data, not the data itself, which saves memory."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "11aeb238-e713-4d49-9bbe-e036e8bf0b68",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds_in = xr.open_dataset('ds_output_file.nc')\n",
- "\n",
- "ds_in"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "fd93ee6f-8f5c-40a1-9ef9-db9b05b93f0f",
- "metadata": {},
- "source": [
- "If you want to load the dataset into memory use load()."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "f5f2911f-b109-4ca3-a590-1fd214fac009",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds_in2 = xr.open_dataset('./ds_output_file.nc').load()"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "2785706a-a822-4998-b91d-93b4cf4184c1",
- "metadata": {},
- "source": [
- "Delete this duplicate dataset."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "f2cd578f-2f45-459a-ac8d-59c8aff4da4f",
- "metadata": {},
- "outputs": [],
- "source": [
- "del(ds_in2)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "72e4b757-e417-4180-b8fe-bdd333af3dba",
- "metadata": {},
- "source": [
- "Read another netCDF file."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "91ae1efb-a7b0-4639-8d18-31dbdb90a785",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds = xr.open_dataset('../data/tsurf.nc')\n",
- "ds.info()"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "2424d12e-3433-42f2-9d39-9edd24d23290",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "## Read a GRIB file\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "c496f4c5-f38e-44a9-80f1-ffc8ba003fce",
- "metadata": {},
- "source": [
- "Now, we can use again the xr.open_dataset() function but this time with the engine 'cfgrib'."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "61c46950-2577-408d-a015-936a7dd618cd",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds2 = xr.open_dataset('../data/MET9_IR108_cosmode_0909210000.grb2',\n",
- " engine='cfgrib')"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "d11b846a-c077-4afa-ac08-8208bf8d9cfc",
- "metadata": {},
- "outputs": [],
- "source": [
- "ds2.variables"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "f3d929c8-11d6-49ec-b602-b741b1be2484",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "<a class=\"anchor\" id=\"read-multi\"></a>\n",
- "## Read multiple files at once\n",
- "\n",
- "Sometimes you get data stored in multiple separate files but you want to have it available in only one Dataset.\n",
- "\n",
- "In the course directory **data** are 3 example files _precip_day01.nc, precip_day02.nc, and precip_day03.nc_, each containing the data of one day in 6 hour intervals. \n",
- "\n",
- "**Xarray** provides the function `xr.open_mfdataset()` to read multiple files in one step as a single dataset. Before you can use `xr.open_mfdataset` make sure that the Python module **dask** is installed in your environment.\n",
- "\n",
- "<br>\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "3df5e0c2-5050-4ae0-949b-7b2c7d7ae58e",
- "metadata": {},
- "outputs": [],
- "source": [
- "!ls -la ../data"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "67620840-af14-4c7b-a1d3-64fb74785186",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "One reason why **Xarray** is very fast with multiple files is that it does not **load** the data when the files are opened. This is possible by using an underlying library named `dask`. You can recognize that by checking for the `precip` variable in `dsm`.\n",
- "\n",
- "First, we open the multiple files precip_day*.nc in the data directory."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "244cf581-6d30-4d0f-9ac4-f5d176187cee",
- "metadata": {},
- "outputs": [],
- "source": [
- "dsm = xr.open_mfdataset('../data/precip_day*.nc')\n",
- "\n",
- "dsm"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "dbaf788a-2643-4552-bed8-4084044f5c4a",
- "metadata": {},
- "outputs": [],
- "source": [
- "dsm.precip[1,4,5]"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "791d6aad-81e2-4029-88b5-57557b88de96",
- "metadata": {},
- "source": [
- "will not show you an exact value but only a description of what this output will be. You would have to load the data into memory first for accessing one specific point of the array. This is most often not necessary for your workflow.\n",
- "\n",
- "The entire array can be loaded into memory by `dsm.precip.load()`. You can also do: \n",
- "```python\n",
- "dsm.precip.values[1,4,5]\n",
- "```\n",
- "\n",
- "âž¡ï¸ While data is not in loaded, you can work on files that are **larger than memory**."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "ff5366f7-3dc5-4421-86e5-7af03a79eae4",
- "metadata": {},
- "outputs": [],
- "source": [
- "dsm.precip[1,4,5]"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "74da3f4d-efa8-404a-8835-a34acb65ac04",
- "metadata": {},
- "outputs": [],
- "source": [
- "dsm.precip.load()"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "828591a2-9cbb-469a-9d30-43eb5b990841",
- "metadata": {},
- "outputs": [],
- "source": [
- "dsm.precip[1,4,5]\n",
- "\n",
- "# is the same as\n",
- "\n",
- "dsm.precip.values[1,4,5]"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "aac42f5a-dc8e-4aff-8175-8b503e9d0887",
- "metadata": {},
- "source": [
- "The `xr.open_mfdataset` function is very powerful. It contains over 10 arguments which allow users to configure how the files are combined:\n",
- "\n",
- "- On what dimension should the data be concatted\n",
- "- How strict should tests ensure that the data can be concatted\n",
- "- What are coordinates, what are data variables"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "e4e748a6-d2f7-49ec-8482-1d31252fa204",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "## Exercise:\n",
- "\n",
- "1. Read the file '../data/rectilinear_grid_2D.nc'\n",
- "2. Print the file content"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "300aba36-5fea-4a1e-bf0a-cbb794cbe21e",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "fb7394f4-6048-4813-a539-7032735be0c7",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 2.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "60b1ce25-8fcb-49b4-a3a1-758b174363ad",
- "metadata": {
- "tags": []
- },
- "source": [
- "<br>\n",
- "\n",
- "### Solution"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "e87e1214-104e-42ae-9d1d-98808a2e2270",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 1.\n",
- "ds_reclin = xr.open_dataset('../data/rectilinear_grid_2D.nc')"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "494d4428-e483-4b9a-b01a-4327f64f1813",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 2.\n",
- "ds_reclin"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "5fc3b4be",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "<br>\n",
- "\n",
- "# Get variable coordinates and names\n",
- "\n",
- "It is always good to have a closer look at the data, and this can be done very easily using the attributes, dimensions, and coordinates explained above.\n",
- "\n",
- "Show the coordinates stored in file:\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "fc09aa0d-d764-4179-b1b8-a3f2eb10d9a5",
- "metadata": {},
- "outputs": [],
- "source": [
- "coords = ds.coords\n",
- "coords"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "8843d365-22c6-45b3-891a-a8c51ed6988c",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "List the variables stored in the file:"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "fedae0e1-40f8-45f6-bbe9-699fb636dee7",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "outputs": [],
- "source": [
- "variables = ds.variables\n",
- "variables"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "fb18921d-de38-466f-bbcd-5f2865d69393",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "Here we can see the time displayed in a readable way, because Xarray use the datetime64 module under the hood. Also the variable and coordinate attributes are displayed."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "ddd3a653",
- "metadata": {
- "slideshow": {
- "slide_type": "subslide"
- }
- },
- "source": [
- "## Exercise\n",
- "\n",
- "Use the Dataset ds from above.\n",
- "\n",
- "1. Print the global file attributes\n",
- "2. What is the difference of list(ds.keys()), list(ds.data_vars), and list(ds) ?\n",
- "3. Print the attributes of the variable of ds\n",
- "\n",
- "\n",
- "<br />"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "1810c8b8-43bc-4e01-9cf6-d75d1a5c291c",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "a7355519-d784-41ee-ba18-712d5981ca5e",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 2.\n"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "407473da-4862-4988-b3aa-ff7120188197",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 3.\n"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "cb3854ee-f667-479d-9e6a-14b88dffaf58",
- "metadata": {},
- "source": [
- "<br>\n",
- "\n",
- "### Solution"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "8cd6f10c-cadf-4b24-9d1d-212736d5c8b9",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 1.\n",
- "print(ds.attrs)\n",
- "\n",
- "print('------------------------------------------------')"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "e9dbc368-6540-4345-9fe4-e19663bf9b5e",
- "metadata": {},
- "outputs": [],
- "source": [
- "# 2.\n",
- "print(list(ds.keys()))\n",
- "print(list(ds.data_vars))\n",
- "print(list(ds))\n",
- "\n",
- "print('------------------------------------------------')"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "f9dfb350-44d3-46d3-8bca-88b665193219",
- "metadata": {
- "tags": []
- },
- "outputs": [],
- "source": [
- "# 3. \n",
- "print(ds.tsurf.attrs)\n",
- "print(ds['tsurf'].attrs)"
- ]
- }
- ],
- "metadata": {
- "kernelspec": {
- "display_name": "pythoncoursekernel",
- "language": "python",
- "name": "pythoncoursekernel"
- },
- "language_info": {
- "codemirror_mode": {
- "name": "ipython",
- "version": 3
- },
- "file_extension": ".py",
- "mimetype": "text/x-python",
- "name": "python",
- "nbconvert_exporter": "python",
- "pygments_lexer": "ipython3",
- "version": "3.12.2"
- },
- "toc-autonumbering": true
- },
- "nbformat": 4,
- "nbformat_minor": 5
-}